Advancing Your Research
Brian Strahl lab, Dept. of Biochemistry and Biophysics
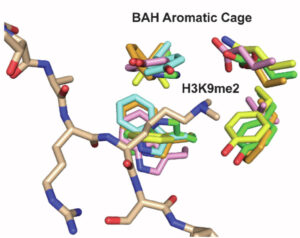
The Strahl lab focuses on the role of histone post-translational modifications, leveraging multiple model systems including mammalian cells and the budding yeast Saccharomyces cerevisiae to understand how histone modifications function. The Strahl lab, in collaboration with the R. L. Juliano Structural Bioinformatics Core, is actively engaged in unraveling the role of the bromo-adjacent homology (BAH) domains, particularly within the context of PBRM1 (BAF180), a key component of the PBAF chromatin-remodeling complex.
Sharon Campbell lab, Dept. of Biochemistry and Biophysics
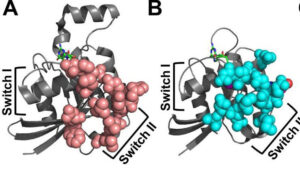
The laboratory of Dr. Campbell employs a multidisciplinary approach focusing on molecular and cellular biology to conduct cell-based adhesion, motility, signaling, and transformation analyses. R. L. Juliano Structural Bioinformatics Core provide computational support, molecular modeling, and Molecular Dynamics simulations to complement experimental data, enhancing our understanding of complex biological processes and aiding in the interpretation of results. Through our integrated approach and collaborative efforts, we aim to advance our understanding of fundamental biological processes and develop novel therapeutic strategies.
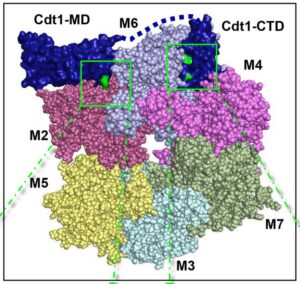
Jean Cook lab, Dept. of Biochemistry and Biophysics
The Cook lab focuses on understanding the eukaryotic cell cycle, with a specific emphasis on unraveling the mechanisms underlying the function of the Cdc10-dependent transcript 1 (Cdt1) protein. Cdt1 is essential for minichromosome maintenance (MCM) loading during the G1 phase of the cell cycle, yet its precise mechanism of action remains incompletely understood. Leveraging experimental approaches in combination with computational methods, the lab aims to elucidate the intricate molecular mechanisms governing Cdt1 function. To support these computational efforts, the lab collaborates closely with the R. L. Juliano Structural Bioinformatics Core, which provides molecular modeling and Molecular Dynamics simulations tailored to investigate the structural dynamics of Cdt1 and its interactions within the cell cycle machinery. This collaborative endeavor between the Cook lab and the Structural Bioinformatics Core is essential for advancing our understanding of the eukaryotic cell cycle and uncovering novel insights into the regulation of cellular processes crucial for cell division and proliferation.
Bob Duronio lab, Integrative Program for Biological and Genome Sciences (IBGS)
The Duronio Lab studies the genetic and epigenetic control of cell cycle progression during animal development. To support computational efforts, the lab benefits from the computational resources and expertise provided by the R. L. Juliano Structural Bioinformatics Core. Through molecular modeling and Molecular Dynamics simulations offered by the Core, the lab aims to gain deeper insights into histone modifications and their functional implications in chromatin biology and epigenetics. This collaborative effort between the Duronio lab and the Structural Bioinformatics Core contributes to advancing our understanding of epigenetic mechanisms and their role in cellular processes.
Greg Matera lab, Integrative Program for Biological and Genome Sciences (IBGS)
The Matera lab focuses on three main research areas: epigenetic gene regulation, animal models of Spinal Muscular Atrophy (SMA), and RNA processing and Ribonucleoprotein (RNP) assembly. In the realm of epigenetic gene regulation, the lab investigates the molecular mechanisms governing the dynamic regulation of gene expression through chromatin modifications and regulatory RNAs. To support computational efforts in understanding the stability of the Survival Motor Neuron (SMN) protein, a key player in SMA pathology, the lab collaborates with the R. L. Juliano Structural Bioinformatics Core. Leveraging computational modeling techniques such as AlphaFold, the Core aids in deciphering the structural dynamics of the SMN protein, providing valuable insights into its stability and function. This collaborative endeavor between the Matera lab and the RLJSBI Core is instrumental in advancing our understanding of epigenetic regulation, neurodegenerative diseases, and RNA biology.

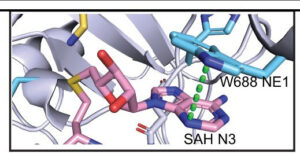
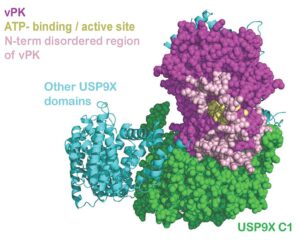
Damania lab, of Microbiology and Immunology
The Damania lab is dedicated to studying the intricate mechanisms by which viruses cause cancer and evade host immunity. With a focus on Kaposi’s sarcoma-associated herpesvirus (KSHV), the lab investigates the molecular pathways underlying viral oncogenesis and immune evasion strategies. In collaboration with the R. L. Juliano Structural Bioinformatics Core, the lab has made significant strides in understanding KSHV pathogenesis. Specifically, through collaborative efforts, they have delineated how KSHV Viral Protein Kinase interacts with USP9X to modulate the viral lifecycle, shedding light on critical interactions that drive viral replication and persistence. This collaboration highlights the essential role of the Structural Bioinformatics Core in advancing our understanding of viral-associated cancers and paving the way for the development of targeted therapies.
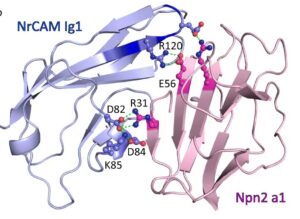
Patricia Maness lab, of Biochemistry and Biophysics
The Maness lab focuses on elucidating the mechanisms underlying the specification of neuronal connectivity by cell recognition molecules. The lab investigates the intricate molecular interactions and signaling pathways mediated by these cell recognition molecules to understand their roles in neural development and function. In collaboration with the R. L. Juliano Structural Bioinformatics Core, the lab has employed AlphaFold to unravel the structural interactions of NrCAM in the Sema3F holoreceptor complex. By deciphering the structural dynamics and binding interfaces of NrCAM, this collaborative effort sheds light on the molecular mechanisms underlying neuronal connectivity and provides valuable insights into the pathogenesis of neurological disorders.
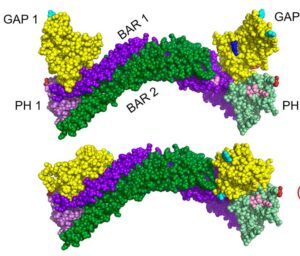
Joan Taylor lab, Department of Pathology and Laboratory Medicine
The Taylor Lab is dedicated to characterizing the intracellular signaling pathways that regulate normal and aberrant growth responses in the cardiovascular and musculoskeletal systems. With a focus on understanding the molecular mechanisms underlying cellular growth and differentiation, the lab investigates key signaling pathways involved in cardiovascular and musculoskeletal development and diseases. In collaboration with the R. L. Juliano Structural Bioinformatics Core, the lab has investigated the BAR-PH mediated autoinhibition of GRAF3. By employing advanced computational modeling techniques, the Structural Bioinformatics Core has provided valuable insights into the structural dynamics and regulatory mechanisms of GRAF3, shedding light on its role in cellular signaling and disease pathogenesis.