Services and Rates
Please, download and complete the Contact Information Form and the specific form for the requested service, and bring them along with your samples.
High Throughput Sequencing
16S rRNA Amplicon Sequencing
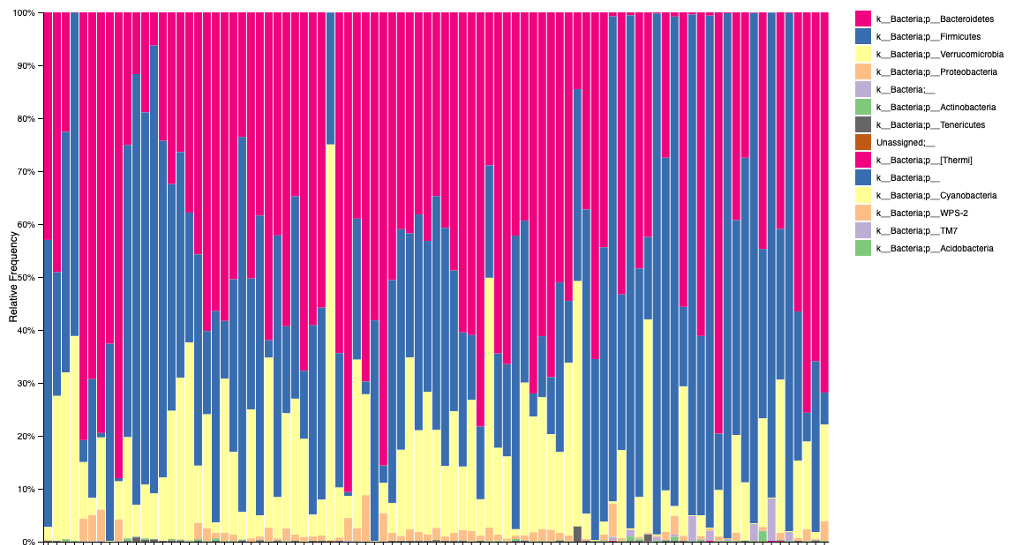
Available Instruments
Quantitative (q)-PCR
Standard qPCR
The BioMark™ Systems paired with the Fluidigm Dynamic Arrays are an efficient solution for large-scale, real-time qPCR. The nanofluidic chips contain fluidic networks that automatically combine sets of samples with sets of assays. This innovative solution for real-time qPCR provides reaction densities far beyond what is possible with microtiter plates and significantly reduces the number of liquid-handling steps and the volume per reaction.
The chip used in this system contains 20,000 individual wells and works by partitioning a standard PCR reaction into thousands of individual PCR reactions. A portion of these partitions contains the target molecule, while other partitions do not, leading to positive and negative reactions. Following amplification, the fraction of negative reactions is used to quantify the number of target molecules in the sample, all without reference to standards or controls.
We offer digital PCR service using QuantStudio 3D™ Digital PCR system from ThermoFisher Scientific. This is a chip based digital PCR platform. In contrast with droplet digital PCR the reaction is taking place in a well of the chip and not in a droplet, with fewer pipetting steps, which decreases the likelihood of cross-contamination.
Applications:
- Absolute quantification of DNA molecules
- Pathogen detection and load determination
- GMO detection and contamination assessment
- Library quantification for next-generation sequencing
- Characterization of low-fold changes in mRNA and miRNA expression
- Rare cancer mutation quantification
- CNV detection
Advantages:
- Digital PCR does not rely on Ct values to quantify copy number, thus, PCR efficiency differences among biological samples are much less likely to affect quantitative results.
- Digital PCR does not rely on standard curve for absolute quantification.
- Digital PCR is much less sensitive to PCR inhibitors present in crude samples.
Consulting and Research Support
Including:
- Sample genomic DNA isolation
- Plasmid and virus nucleic acids isolation
- RNA isolation
- Strain typing
- Optimization of bacterial culture conditions
- High-throughput liquid handling (PCR reaction set up, sample pooling, picogreen quantification of nucleic acids
Use the link to book an appointment for consultation and research support.
AMC Culture Collection
The understanding of host-associated and free-living microbial communities’ structure and functionality has greatly advanced via high-throughput sequencing. As the field moves from association studies to mechanistic and interventional studies, the availability of authenticated, reliable biological material and associated information has become crucial.In partnership with the Center for Gastrointestinal Biology and Disease (CGIBD) Advanced Analytics Core (AAC) and the Carolina Donor Services (CDS), the federally designated organ procurement organization serving 7.2 million people in 77 counties of North Carolina and Virginia, the Microbiome Core has isolated, characterized and preserved over 500 human isolates and reference probiotic bacteria of medical and scientific relevance.
The AMC Culture Collection adds to our current services of microbiome analysis to provide well-characterized, active and viable strains for microbiome studies that demand specific bacterial groups and/or functionality. Contact us at microbiome@med.unc.edu for more information or a consultation.
New Services:
- Strain/consortium request: This service provides well-characterized human isolates and reference probiotic bacteria for research. Lyophilized, physiologically active or glycerol stocks can be provided as requested.
- Strain submission for storage and preservation: Researchers can submit relevant strains for long term storage and preservation. Service can include strain characterization (antibiotic resistance, carbohydrate utilization and genome sequencing).
- Activation and culturing of strains: The service includes the activation of bacterial cultures under physiologically appropriate conditions. The Core will provide cultures in liquid or solid media as requested.


