Single Cell Solutions (10x Genomics)
10x Genomics Single Cell Services:
- FACS
- Single cell library preparation (RNA-Seq, ATAC-Seq, etc…)
- Sequencing and basic data processing
The Advanced Analytics Core now offers turnkey 10x Genomics single cell services. Simply provide a single cell suspension, and the Core will perform the services necessary to generate high quality sequencing data: cell sorting, library preparation, sequencing, and basic data analysis.
The 10x Genomics Chromium System encapsulates hundreds to tens of thousands of cells (or nuclei) into uniquely addressable partitions in minutes, each containing an identifying barcode for downstream analysis. Up to eight samples can be processed simultaneously, and multiple runs can be conducted in a single day. Encapsulation is not biased by size and is compatible with fresh, frozen, and methanol-fixed samples. Applications of the Chromium System include:
- Gene expression profiling
- Immune profiling
- CRISPR screening
- Copy number variation
- Chromatin accessibility (ATAC-Seq)
- Cell surface protein characterization (CITE-Seq/REAP-Seq)
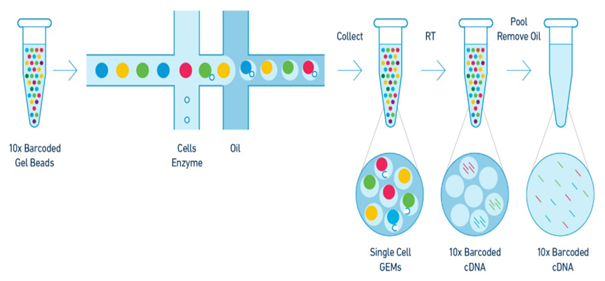
GemCode Technology provides a precisely engineered reagent delivery method that enables thousands of micro-reactions in parallel. Gel beads infused with millions of barcoded oligonucleotides are mixed with a sample and then added to an oil-surfactant solution to create gel beads in emulsion (GEMs). Barcoded products are pooled for downstream reactions to create short-read sequencer compatible libraries. After sequencing, the resulting barcoded sequences are fed into analysis pipelines that use the barcode information to map reads back to their original cell.
Instruments:
10x Genomics Chromium Single Cell Controller
Services:
FACS: Sample preparation via FACS can be completed by the Core. Contact the Associate Director to arrange a consultation.
Single-Cell Isolation & 10x Library Preparation: Each 10x chip is comprised of eight sample inlets into which single cell suspensions are loaded. Each inlet can encapsulate as many as 10,000 cells or as few as 500 cells. Multiple chips can be run on a single day. The Core recommends providing at least 50,000 cells in suspension per sample, as fewer cells can make important QC steps difficult to accomplish.
Other 10x protocols will include different deliverables. Please contact the r for more information.
How to start a project:
To begin a project using the 10x system, please contact the Genomic Services Manager to arrange a consultation. The AA Core will then schedule the project, coordinate with the end-user for cell drop off, and discuss pricing and turnaround estimates for project completion.
Pricing estimates:
10x Single Cell 3’ or 5’ Library Preparation and QC
| Number of 10x Samples | Price per Sample* | UNC/NCSU/CGIBD Member Price** |
| First Sample | ~$3,673.18 | ~$3,173.50 |
| Samples 2 through 8 | ~$2,228.28 | ~$2,145.00 |
Pricing for other 10x single cell services is available upon request: Immune profiling, CRISPR screening, copy number variation, cell surface protein characterization (CITE-Seq/REAP-Seq). Please contact the Genomic Services Manager for a personalized quote.
** Investigators affiliated with UNC, NCSU, or the CGIBD receive discounted pricing.
Submitting Samples:
Users may schedule an appointment to drop off samples in the AA Core single cell laboratory:
AA Core MBRB Laboratory
Medical Biomolecular Research Building (MBRB)
Room 4331
111 Mason Farm Rd
Chapel Hill, NC 27599