Recent Publications
Harlin Lee, Boyue Li, Shelly DeForte, Mark L. Splaingard, Yungui Huang, Yuejie Chi & Simon L. Linwood, published in Nature: A large collection of real-world pediatric sleep studies
William H. Weir, Peter J. Mucha, William Y. Kim, published in Cell: A bipartite graph-based expected networks approach identifies DDR genes not associated with TMB yet predictive of immune checkpoint blockade response 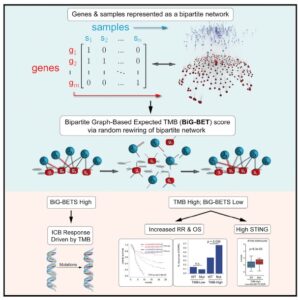
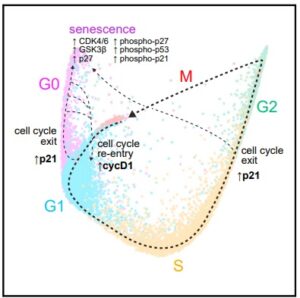
Wayne Stallaert, Ph.D., Katarzyna M. Kedziora, et al., published in Cell Systems: The structure of the human cell cycle.
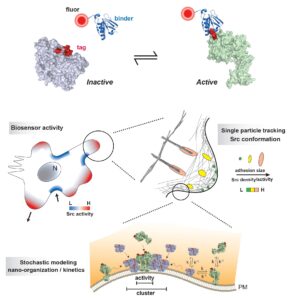
Bei Liu, Orrin J. Stone, Michael Pablo, Cody Herron, Ana T. Nogueira, Onur Dagliyan, Jonathan B. Grimm, Luke D. Lavis, Timothy C. Elston, Klaus M. Hahn, publication in Cell: Biosensors based on peptide exposure show single molecule conformations in live cells
 Haidong Yi, Natalie Stanley, publication in bioRxiv: CytoSet: Predicting clinical outcomes via set-modeling of cytometry data
Haidong Yi, Natalie Stanley, publication in bioRxiv: CytoSet: Predicting clinical outcomes via set-modeling of cytometry data
 Khem Raj Ghusinga publication in PNAS: Molecular switch architecture determines response properties of signaling pathways.
Khem Raj Ghusinga publication in PNAS: Molecular switch architecture determines response properties of signaling pathways.
 Amy Pomeroy publication in Science Signaling: A predictive model of gene expression reveals the role of network motifs in the mating response of yeast.
Amy Pomeroy publication in Science Signaling: A predictive model of gene expression reveals the role of network motifs in the mating response of yeast.
 Rau Lab publication in PLoS Genetics: Modeling epistasis in mice and yeast using the proportion of two or more distinct genetic backgrounds: Evidence for “polygenic epistasis”.
Rau Lab publication in PLoS Genetics: Modeling epistasis in mice and yeast using the proportion of two or more distinct genetic backgrounds: Evidence for “polygenic epistasis”.

Adam Palmer’s publication in Cancer Research: A Proof of Concept for Biomarker-Guided Targeted Therapy against Ovarian Cancer Based on Patient-Derived Tumor Xenografts.
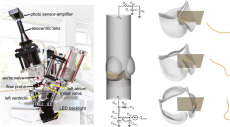 Mathematics Department’s publication in Elsevier: Bioprosthetic aortic valve diameter and thickness are directly related to leaflet fluttering: Results from a combined experimental and computational modeling study.
Mathematics Department’s publication in Elsevier: Bioprosthetic aortic valve diameter and thickness are directly related to leaflet fluttering: Results from a combined experimental and computational modeling study.
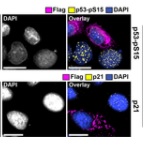 Labs: Dittmer, Damania, Baric, Heise and U. Wisconsin’s Gabriele Neumann, Peter Halfmann, Yoshihiro Kawaoka’s publication of Novel modulators of p53-signaling encoded by unknown genes of emerging viruses.
Labs: Dittmer, Damania, Baric, Heise and U. Wisconsin’s Gabriele Neumann, Peter Halfmann, Yoshihiro Kawaoka’s publication of Novel modulators of p53-signaling encoded by unknown genes of emerging viruses.
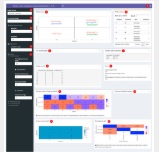
Michael Love Lab published in F1000 Research ExploreModelMatrix: Interactive exploration for improved understanding of design matrices and linear models in R [version 2; peer review: 3 approved]

Amy Shaub Maddox Lab posted a publication in bioRxiv: Novel cytokinetic ring components limit RhoA activity and contractility.
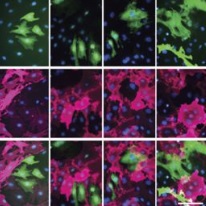
Qian Lab’s recent publication in Nature: Single-cell transcriptomic reconstructs fate conversion from fibroblast to cardiomyocyte.

The recent publication using genomic and computational approaches to identify and annotate non-coding RNAs transcribed at enhancer regions known as enhancer RNAs in breast cancer cells: Enhancer transcription revealed subtype-specific gene expression programs controlling breast cancer pathogenesis.
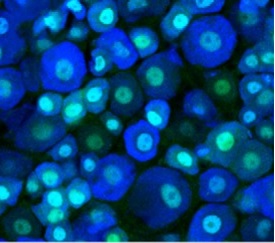
Zylka lab, studies genetic and environmental risks for autism. In addition, Zylka lab study molecular and brain mechanisms that underlie pain sensation. Their long-term goal is to uncover new treatments for chronic pain and neurodevelopmental disorders, like autism, Rett syndrome and Angelman syndrome. A collection of the publications can be found on the NIH website.
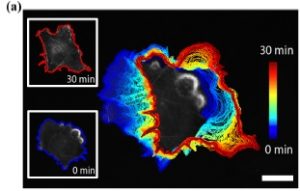
Many studies employ shape descriptors globally to probe mechanisms of cell morphogenesis. Here, we examined cell behavior very locally along the cell edge. The Hilbert-Huang transform (HHT) was used to extract spectra of instantaneous edge motion frequency and magnitude along the entire cell edge, and these were then used to classify subcellular edge sectors with distinct dynamics. When optogenetics was used to acutely inhibit specific signaling pathways, we found changes in the frequency spectra, but not in the magnitude spectra. After clustering cell edge sectors with distinct morphodynamics we observed that sectors with different frequency spectra are associated with specific signaling dynamics and motility behavior. Together these observations let us conclude that the frequency spectrum encodes the wiring of the molecular circuitry that regulates edge movements, whereas the magnitude captures the activation level of circuitry. Full results can be found in the publication: Profiling cellular morphodynamics by spatiotemporal spectrum decomposition.
Calabrese Lab developed a new method to quantify sequence similarity between evolutionarily unrelated lncRNAs. The method allows us to take sequence information from a well-studied lncRNA, and use it to discover lncRNAs that may be functioning through a related mechanism. Full results can be found in the publication: Functional classification of long non-coding RNAs by k-mer content.
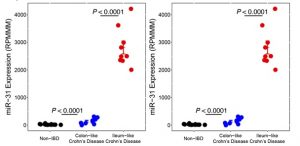
MicroRNA profiling in the colon from Crohn’s disease (CD) patients revealed two distinct molecular subtypes, with miR-31 the most significantly differentially expressed between the two. Low colonic miR-31 expression levels at the time of surgery were associated with worse disease outcome in adults and with future development of fibrostenotic ileal CD requiring surgery in children. Full results can be found in the publication: Colonic epithelial miR-31 associates with the development of Crohn’s phenotypes.
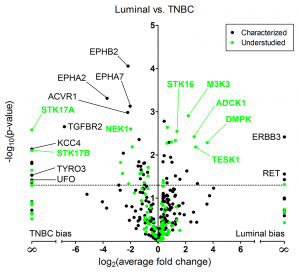 Recent work by the Gomez and Johnson labs has led to the discovery of multiple protein kinases and related kinome subnetworks that have distinctive behaviors across breast cancer subtypes as well as in response to drug treatment. Of note was the identification of a number of poorly understood or “dark” kinases that had highly dynamic behavior and thus may represent valuable targets for future drug development. Full results can be found in the publication: Proteomic analysis defines kinase taxonomies specific for subtypes of breast cancer
Recent work by the Gomez and Johnson labs has led to the discovery of multiple protein kinases and related kinome subnetworks that have distinctive behaviors across breast cancer subtypes as well as in response to drug treatment. Of note was the identification of a number of poorly understood or “dark” kinases that had highly dynamic behavior and thus may represent valuable targets for future drug development. Full results can be found in the publication: Proteomic analysis defines kinase taxonomies specific for subtypes of breast cancer