Sunday, June 27, 2010 — In a paper published in the journal Nature Biotechnology, UNC’s Klaus M. Hahn, PhD, describes a new technique called engineered allosteric regulation, which provides a new tool for scientists who study the interactions of proteins within living cells.

on the paper published in Nature Biotechnology this week.
In a paper published and highlighted today in the journal Nature Biotechnology, Klaus Hahn, PhD, who is the Thurman Professor of Pharmacology at the University of North Carolina at Chapel Hill and a member of UNC Lineberger Comprehensive Cancer Center, describes a new technique called engineered allosteric regulation, which provides a new tool for scientists who study the interactions of proteins within living cells.
“Engineered allosteric regulation is a new method that provides precise control of kinase activity in living cells,” said Hahn.
“We can now take the kinase of choice and precisely control the ‘on/off’ switch, thereby seeing what they are doing and how they control cell function. The technology has exciting applications in basic research, since kinases are the central regulators of almost every cellular process. The ability to precisely control the state and timing of kinase action within cells opens the door to a broad range of new scientific insights,” he added.
Being able to turn a specific kinase off and on creates a powerful tool for researchers in understanding the specific role each kinase plays in cell signaling pathways within cells. Understanding the role of kinase and how they specifically control cellular processes is particularly helpful to understanding not only normal cellular processes and development but also the development of diseases such as cancer.
First author on the paper is Andrei V. Karginov, of the Department of Pharmacology. Other contributing authors from the UNC Department of Biochemistry and Biophysics are Feng Ding, Pradeep Kota and Nikolay V. Dokholyan.
The research for this paper was supported by the UNC Cancer Research Fund and the National Institutes of Health for funding (GM64346 and GM057464 to K.M.H.; GM080742 and GM080742- 03S1 to N.V.D.).
Article in School of Medicine news
Link to full text article in Nature Biotechnology
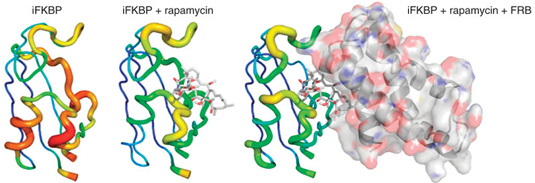
Changes in the molecular dynamics of iFKBP upon binding to rapamycin and FRB. Warmer colors and thicker backbone indicate increasing root mean square fluctuation. From Fig. 1: Design and generation of RapR-FAK.
