Congratulations to Blossom Damania, PhD and her lab on the publication in Cell Reports of a joint paper on “High-density amplicon sequencing identifies community spread and ongoing evolution of SARS-CoV-2 in the Southern United States.” Danielle Chappell, PHCO graduate student and Damania lab member, is a co-author. Dirk Dittmer, Melissa Miller and their labs collaborated on the project.
Summary/Abstract
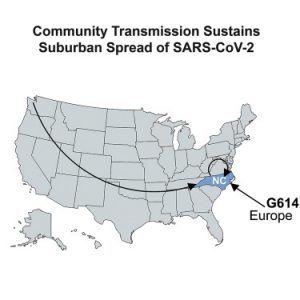
SARS-CoV-2 is constantly evolving. Prior studies focused on high case-density locations, such as the U.S. Northern and Western metropolitan areas. This study demonstrates continued SARS-CoV-2 evolution in a suburban Southern U.S. region by high-density amplicon sequencing of symptomatic cases. 57% of strains carried the spike D614G variant, which was associated with higher genome copy numbers and its prevalence expanded with time. Four strains carried a deletion in a predicted stem loop of the 3’ untranslated region. The data are consistent with community spread within local populations and the larger continental U.S. The data instill confidence in current testing sensitivity and validate “testing by sequencing” as an option to uncover cases, particularly non-standard COVID-19 clinical presentations. This study contributes to the understanding of COVID-19 through an extensive set of genomes from a non-urban setting and informs vaccine design by defining D614G as a dominant and emergent SARS-CoV-2 isolate in the U.S.
Blossom Damania, PhD, is a Professor and Vice Dean of Research in the Department of Microbiology & Immunology, and a joint Professor in the Department of Pharmacology. Collaborators on the research project are Dirk Dittmer, PhD, lead author on the paper and Professor in the Department of Microbiology & Immunology, Melissa Miller, PhD, Professor in the Department of Pathology & Laboratory Medicine, and their labs.
PHCO graduate student and member of the Damania Lab, Danielle Chappell, is a co-author on this landmark study on the evolution of the virus that causes COVID-19. Congratulations, Danielle!