The Human Pluripotent Cell Core expertise is available to facilitate the development of joint proposals, provide technical support and consultation for researchers working with stem cells.
The Human Pluripotent Cell Core Facility strives to provide:
CUTTING EDGE TECHNOLOGY applied to cell programming, gene editing and cell differentiation.
EXPERTISE in experimental design and development of new methods, and
REPRODUCIBILITY in the results by using standardized methods and well design experiments.
Services include:
- Gene editing of hES, iPSC, stem and cancer cells using CRISPR/ cas9 to create gene knock-out and knock-ins for isogenic controls, as well as reporter lines.
- Reprogramming of blood, fibroblast and other somatic cells into iPSCs. Including iPSC characterization and karyotyping.
- Human hES and iPSC directed differentiation into a variety of cell types in monolayer
- Organoids.
- Method and assay development.
Core Highlight: Tagging endogenous genes in hES, iPSC, stem and cancer cells using CRIPSR/cas9 system.
Live-cell reporters allow the study of protein dynamics in real time and provide clues about their protein localization, function and regulation to better understand the regulatory mechanisms of biological systems.
The HPSCC core provide tagging of endogenous genes in hES, iPSC, stem and cancer cells using CRIPSR/cas9 system. The use of these reporters are unlimited, for example:
- Tagged iPSC cells differentiated into specific cell types can provide mechanistic information of different biological systems to better understand both health and disease.
- Tagged cancer cells can be used to study specific disease mechanism or for drug discover efforts.
Our expertise is available to facilitate the development of joint proposals, provide technical support and consultation for researchers working with stem cells. Please contact the HPSSC Director Adriana S. Beltran, PhD, to discuss your research project.
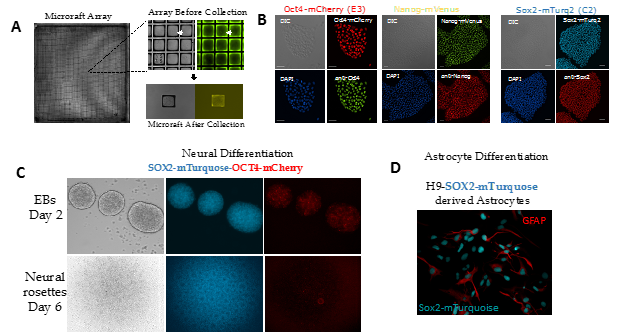
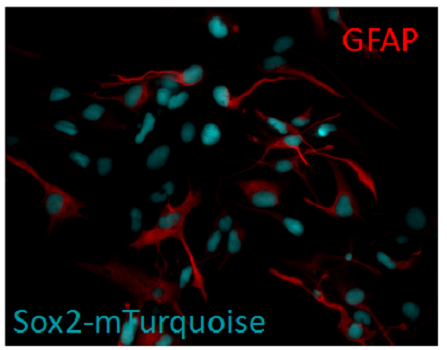
Figure 1. Generation of reporter hESC lines using CRIPSR/Cas9. A) A mixed population of H9 hESCs seeded on a microraft array. The array was scanned via high-content imaging to identify positive clones. Each individual colony was allowed to expand across a microraft and then screened for correct FP insertion. B) Single reporter cell lines are fixed and labeled with a corresponding antibody to determine how well the fluorescent protein tag correlated with total protein levels. C). Double reporter line undergoes ectoderm differentiation in 2D and 3D. C). SOX2-tagged hES cells differentiate into astrocytes.
*Work in collaboration with Sam Wolf and Jeremy Purvis.