The Hooker Imaging Core (HIC) is an open core providing a broad range of light microscopy techniques, analyses and consultation for UNC researchers. The core is located in Taylor Hall near Lineberger and the MBRB and includes several confocal and widefield microscopes in addition to systems for high content screening and long-term live cell imaging. The core also provides users a full tissue culture suite including incubators, hood and refrigerator. Training and other assistance is provided by the core managers. After training, users will independently perform their experimental research with consultation available from the co-directors as needed.
Available Instruments
Leading the instrument lineup are four confocal microscopes including the flagship confocal, a Zeiss 880, the latest model with superb image quality coupled with extraordinary sensitivity and a user-friendly interface. This inverted microscope is also equipped with the Airy scan and fast Airy scan options giving super resolution performance when required. The figure below compares a conventional confocal image (left panel) with an Airy scan image showing improved resolution (right panel). The instrument has an on-stage environmental chamber to support live-cell imaging. Confocal imaging is available at a large number of excitation wavelengths ranging from violet (405 nm) to far red (633 nm).
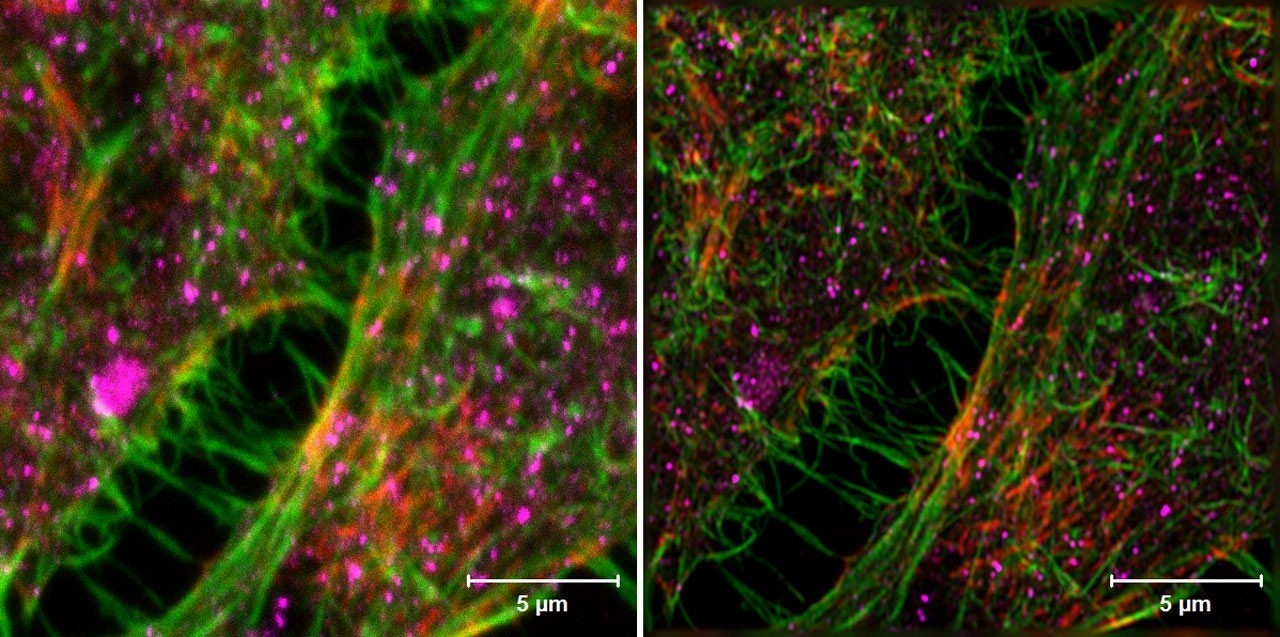
Confocal image (left) compared to Airyscan (right). HeLa cells.Sample: Courtesy of S. Traikov, BIOTEC, TU Dresden, Germany. https://www.zeiss.com
A Zeiss 800 upright confocal microscope is available for fixed samples and wet specimens such as whole Zebrafish. An Olympus FV1000 confocal microscope, completely suitable for fixed and live cell specimens, is now available.
Four widefield microscopes are equipped to provide color and low light level imaging with acquisition speeds of up to 30 frames per second. Applications include ratio imaging for calcium concentration and automated multi-time, multi-position, multi-wavelength, z-stack acquisition of fluorescence images and tiling of multiple images to create a large field of view. One of the widefield microscopes is the fluorescence-equipped Leica dissecting scope for macroscopic specimens.
The HIC now has instruments to monitor the behavior of large numbers of cells over time. An Olympus VivaView microscope is capable of multi-dish monitoring of cell behavior over extended periods in response to environmental influences such as drugs or other stimuli.
In addition, the HIC now features high content screening with the GE IN Cell 2200. The IN Cell is a fast and sensitive wide field imaging system that can be fine-tuned to meet the needs of specific high content imaging workflows. The system is equipped with filters to produce fluorescence images using fluorophores emitting from the blue to far red along with companion transmitted light images. In addition, there is environmental control for time lapse imaging. The IN Cell can accommodate slides or multiwell plates with up to 1536 wells.
Other Services
The HIC provides computer workstations equipped with software for off-line processing of acquired images including both commercial (Volocity, Imaris and Matlab) and open source (ImageJ, CellProfiler, etc.) analysis software. Image recording, processing and analysis support is available.
The HIC now has the capability to assist users in the development of computational models on the Virtual Cell platform which is a unique software platform designed to model cell biological processes. The Virtual Cell is open-source software which was created and is now developed and maintained in the Center for Cell Analysis and Modeling at the University of Connecticut Health Center. The computational model can integrate a variety of molecular mechanisms including reaction kinetics, diffusion, flow, membrane transport, lateral membrane diffusion and electrophysiology, and can associate these mechanisms with geometries derived from experimental microscope images. The computational model also can be based on purely theoretical assumptions and help to evaluate hypotheses and to predict behavior of complex, highly non-linear systems. The figure below schematically describes the types of computational models the Virtual Cell can support. Consultation on employing these models is now available in the HIC.
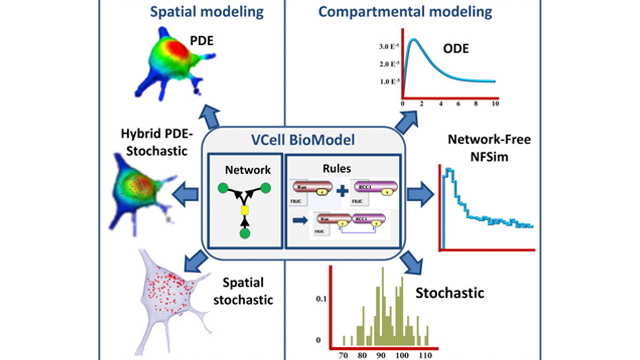
Image credit: Blinov, M.L., et al. (2017). Doi: http://dx.doi.org/10.1016/j.bpj.2017.08.022
The figure shows the types of models that the Virtual Cell can accommodate.
HIC Staff
Core Operations Director: Robert Currin
Core Faculty Advisor: Ken Jacobson, Kenan Professor of Cell Biology and Physiology
Image processing, analysis and modeling consultation is provided by Dr. Maryna Kapustina, Research Associate Professor of Cell Biology and Physiology