Research Interests
DNA damage, DNA replication, DNA repair, signal transduction, genome maintenance, carcinogenesis, cancer therapy
Our overall goal is to elucidate mechanisms by which cells sense, repair and tolerate DNA damage, and to understand how loss of these ‘genome maintenance’ mechanisms leads to disease.
Human cells are frequently exposed to DNA-damaging agents from environmental and endogenous sources. Failure to tolerate DNA damage can lead to loss of cell viability and altered homeostasis (e.g. bone marrow failure syndromes arising from reduced DNA damage tolerance of hematopoietic progenitors). Moreover, failure to replicate and repair damaged DNA accurately can lead to genomic instability and cancer. Indeed many cancer-propensity syndromes result from defective DNA repair.
DNA repair is also very relevant to cancer treatment: radiotherapy and many chemotherapeutic drugs kill cancer cells by causing irreparable DNA damage. Unfortunately, cancer cells can acquire resistance to chemotherapy and radiotherapy via activation of DNA repair pathways. Therefore, DNA repair proteins represent potentially valuable therapeutic targets in the cancer clinic.
Much of our research is focused on a DNA damage tolerance mechanism termed ‘Trans-Lesion Synthesis’ (TLS): In response to many DNA-damaging exposures, specialized TLS DNA polymerases are recruited to sites of DNA damage where they replicate and repair damaged DNA, thereby conferring DNA damage tolerance. However TLS is an inherently error-prone process and must be used sparingly to avoid genome instability. Our laboratory is interested in the signal transduction mechanisms that regulate TLS, promote genome maintenance, and guard against cancer in normal cells. We also seek to identify TLS inhibitors that will improve the effectiveness of DNA-damaging therapeutic agents against chemo-resistant cancer cells.
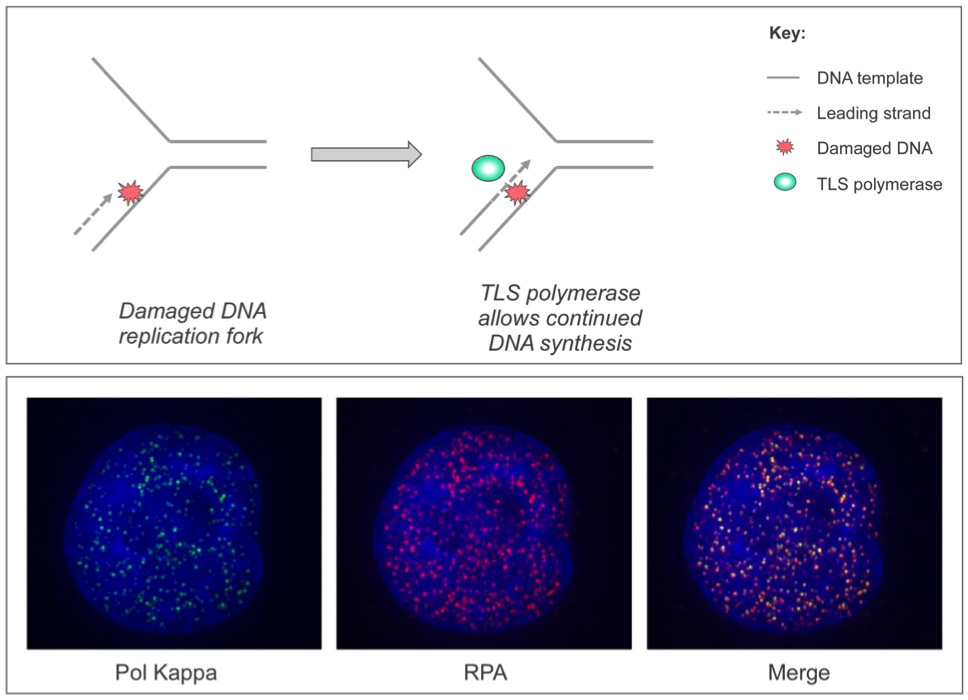
Figure Legend
Upper Panel: When cells acquire DNA damage (red ‘explosion’), specialized TLS DNA polymerases (green sphere) are recruited to DNA replication forks to allow completion of DNA synthesis, thereby conferring DNA damage tolerance. Lower Panel: Confocal microscopy images showing DNA damage-inducible nuclear co-localization of the TLS DNA polymerase Pol Kappa (green foci) with the DNA replication fork protein RPA (red foci).
Selected Publications
Gao Y, Mutter-Rottmayer E, Zlatanou A, Vaziri C, Yang Y. (2017) Mechanisms of Post-Replication DNA Repair. Genes (Basel) 8;8(2). pii: E64. doi: 10.3390/genes8020064.
Mutter-Rottmayer E, Gao Y, Vaziri C. (2016) Cancer cells activate damage-tolerant and error-prone DNA synthesis. Mol Cell Oncol. 3(6):e1225547. doi: 10.1080/23723556.2016.1225547.
Gao Y, Tateishi S., Vaziri C. (2016) Pathological Trans-Lesion Synthesis in Cancer. Cell Cycle 2016 15(22):3005-3006.
Gao Y, Mutter-Rottmayer E, Greenwalt A M, Goldfarb D, Yan F, Yang Y, Martinez RC, Pearce KH, Tateishi S, Major MB, Vaziri C. (2016) A Neomorphic cancer cell-Specific Role of MAGE-A4 in Trans-Lesion Synthesis. Nature Commun. 2016 Jul 5;7:12105. doi: 10.1038/ncomms12105
Yang Y, Poe JC, Yang L, Fedoriw A, Desai S, Magnuson T, Li Z, Fedoriw Y, Araki K, Gao Y, Tateishi S, Sarantopoulos S, Vaziri C. (2015) Rad18 confers hematopoietic progenitor cell DNA damage tolerance independently of the Fanconi Anemia pathway in vivo. Nucleic Acids Res. 2016 44(9):4174-88
Vaziri C, Tateishi S, Yang Y,1 Greenwalt A. (2014) Regulation of Y-Family Translesion Synthesis (TLS) DNA polymerases by RAD18. In ‘Translesion DNA polymerases: from DNA repair and beyond’ Editors: Domenico Maiorano & Dr. Jean-Sébastian Hoffmann (see coming title section of research signpost website http://www.ressign.com/).
Bakkenist CJ, Vaziri C. (2013) Stoichiometry of ubiquitin-binding proteins directs DSB repair. Cell Cycle. 12(24):3716-17.
Durando M, Tateishi S, Vaziri C. (2013) A non-catalytic role of DNA polymerase eta in recruiting Rad18 and promoting PCNA monoubiquitination at stalled replication forks. Nucleic Acids Res. 41(5):3079-93.
Yang Y, Durando M, Smith-Roe SL, Sproul C, Greenwalt AM, Kaufmann W, Oh S, Hendrickson EA, Vaziri C. (2013) Cell cycle stage-specific roles of Rad18 in tolerance and repair of oxidative DNA damage. Nucleic Acids Res. 41(4):2296-312.
Barkley LR, Palle K, Durando M, Day TA, Gurkar A, Kakusho N, Li J, Masai H, Vaziri, C. (2012) c-Jun N-terminal kinase-mediated Rad18 phosphorylation facilitates Pol eta recruitment to stalled replication forks. Mol Biol Cell. 23(10):1943-54.
Williams SA, Longerich S, Sung P, Vaziri C, Kupfer GM. (2011) The E3 ubiquitin ligase RAD18 regulates ubiquitylation and chromatin loading of FANCD2 and FANCI. Blood 117(19):5078-87.
Vaziri C, Masai H. (2010) Integrating DNA Replication with Trans-Lesion Synthesis via Cdc7. Cell Cycle 9(24):4818-23.
Vaziri C. (2010) Linking Cdc7 with the Replication Checkpoint. Cell Cycle 9(24):4787.
Day TA, Palle K, Barkley LR, Kakusho N, Zou Y, Tateishi S, Verreault A, Masai H, Vaziri C. (2010) Phosphorylated Rad18 directs DNA Polymerase eta to sites of stalled replication. J Cell Biol. 191(5):953-66.
Barkley LR, Song IY, Zou Y, Vaziri C. (2009) Reduced Expression of GINS Complex Members Induces Hallmarks of Pre-Malignancy in Primary Untransformed Human Cells. Cell Cycle 8(10):1577-88. (This article was the subject of two separate promotional ‘News & Views’ features).
Liu P, Slater DM, Lenberg M, Nevis K, Cook JG, Vaziri C. (2009) Replication Licensing Promotes Cyclin D1 expression and G1 Progression in Untransformed Human Cells. Cell Cycle 8(1):125-36. (This article was selected by the journal editor for a promotional ‘News & Views’ feature).
Liu P, Barkley LR, Day T, Bi X, Slater DM, Alexandrow MG, Nasheuer HP, Vaziri C. (2006) The Chk1-Mediated S-phase Checkpoint Targets Initiation Factor Cdc45 via a Cdc25A/CDK2-Independent Mechanism. J. Biol. Chem. 281(41):30631-44.
Bi X, Barkley LR, Slater DM, Tateishi S, Yamaizumi M, Ohmori H, Vaziri C. (2006) Rad18 Regulates DNA Polymerase Kappa and is Required for Recovery From S-Phase Checkpoint-Mediated Arrest. Mol. Cell Biol. 26(9):3527-40.
Bi X, Slater DM, Ohmori H, Vaziri C. (2005) DNA Polymerase Kappa is Specifically Required for Recovery from the Benzo[a]Pyrene-Dihydrodiol Epoxide (BPDE)-Induced S-phase Checkpoint. J. Biol Chem. 280(23):22343-55.
Vaziri C, Saxena S, Jeon Y, Lee C, Murata K, Machida Y, Wagle N, Hwang DS, Dutta A. (2003) A p53-Dependent Checkpoint Pathway Prevents Replication. Molecular Cell 11(4):997-1008.
List of publications from PubMed

