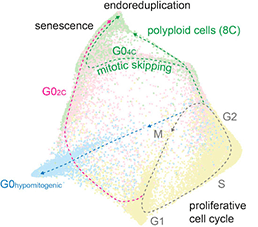
The Architecture of Cell Cycle Arrest
Cell cycle mapping is used to reveal how the cell cycle responds to hypomitogenic, replicative and oxidative stress. Regardless of the phase of cell cycle exit, cells converge on a single state of senescence with a G1-like molecular signature. Stallaert W, Taylor SR, Kedziora KM, Johnson MS, Taylor CD, Sobon HK, Young CL, Limas JC, Varblow-Holloway J, Cook JG, Purvis JE. The molecular architecture of cell cycle arrest (2022). Molecular Systems Biology 18(9):e11087. PMID: 36161508
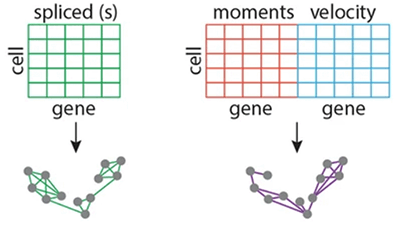
Integrating Temporal Gene Expression Modalities
Integrated data more accurately infers biological trajectories and achieves increased performance on classifying cells according to perturbation and disease states. In many cases, simple concatenation of spliced and unspliced molecules performs consistently well on classification tasks and can be used over more memory intensive and computationally expensive methods. Ranek JS, Stanley N, Purvis JE. Integrating temporal single-cell gene expression modalities for trajectory inference and disease prediction (2022). Genome Biology 23(1):186. PMID: 36064614

Field Trip to the Purvis Lab
Support from the National Science Foundation helped third-grade students from Durham's Central Park School of Children experience single-cell biology. Students learned the basic anatomy of the cell and how information is stored in DNA. They harvested their own cells, examined them under the microscope, and measured image features such as shape, size, and nuclear intensity.
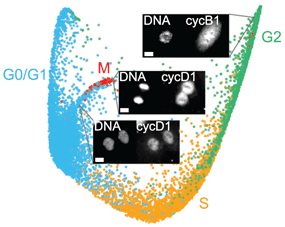
The Structure of the Human Cell Cycle
A complete visualization of the human cell cycle is rendered by combining highly multiplexed single-cell imaging and manifold learning. Stallaert W, Kedziora KM, Taylor CD, Zikry TM, Ranek JS, Sobon HK, Taylor SR, Young CL, Cook JG, Purvis JE. The Structure of the Human Cell Cycle. (2021) Cell Systems Nov 17:S2405-4712(21)00418-X PMID: 34800361
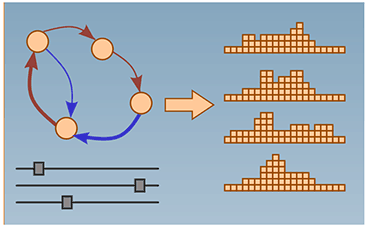
The Cell Cycle Browser
We built an interactive web interface based on real-time reporter data collected in proliferating human cells. This tool facilitates visualizing, organizing, simulating, and predicting the outcomes of perturbing cell-cycle parameters. Borland D, Yi H, Grant GD, Kedziora KM, Chao HX, Haggerty RA, Kumar J, Wolff SC, Cook JG, Purvis JE. The Cell Cycle Browser: An Interactive Tool for Visualizing, Simulating, and Perturbing Cell-Cycle Progression. (2018) Cell Systems 7(2):180-4. PMID: 30077635

DNA Damage Checkpoint Dynamics
Each cell-cycle phase shows a distinct sensitivity and temporal response to DNA damage, explaining why differences in the timing of DNA damage can lead to heterogeneous cell fate outcomes. Chao HX, Poovey CE, Privette AA, Grant GD, Chao HY, Cook JG, Purvis JE. Orchestration of DNA Damage Checkpoint Dynamics across the Human Cell Cycle. (2017) Cell Systems 5(5):445-459.e5. PMID: 29102360