 Welcome to the R. L. Juliano Structural Bioinformatics Core facility at UNC-Chapel Hill. We provide consultations and collaborations on research studies requiring computational structural biology methods. The analyses available through the core are not limited to the study of static structures, but also include molecular dynamics studies for analyzing the contribution of dynamic and collective motions to macromolecular functionality. When experimental structures are not available, molecular modeling studies, whereby the structure of the protein of interest is predicted using known template structures, provide 3D atomic data. All of these analyses contribute to the development of hypotheses to be tested in the laboratory, and give insight into experimental data.
Welcome to the R. L. Juliano Structural Bioinformatics Core facility at UNC-Chapel Hill. We provide consultations and collaborations on research studies requiring computational structural biology methods. The analyses available through the core are not limited to the study of static structures, but also include molecular dynamics studies for analyzing the contribution of dynamic and collective motions to macromolecular functionality. When experimental structures are not available, molecular modeling studies, whereby the structure of the protein of interest is predicted using known template structures, provide 3D atomic data. All of these analyses contribute to the development of hypotheses to be tested in the laboratory, and give insight into experimental data.
- For more information on what services are available, go to: How We Work
- To learn about available software packages, go to: Resources
- For examples of current research using our tools, go to: Advancing Your Research
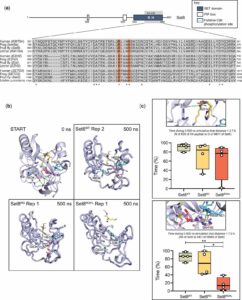
Exploring Key Insights from Diverse MD Simulations and Molecular Modeling Techniques
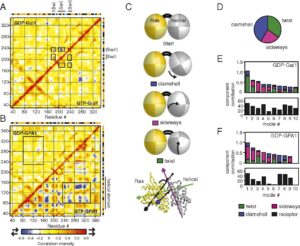
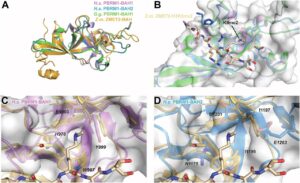
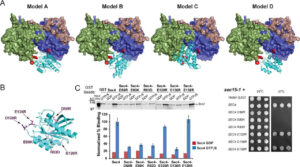
![]()
We obtain on-going support from the UNC Lineberger Comprehensive Cancer Center through the University Cancer Research Fund and the Cancer Center Support Grant. Consequently, publications supported by the UNC Center for Structural Biology must acknowledge NIH grant P30CA016086 and be submitted to PubMed Central in compliance with the NIH Public Access Policy.
Suggested acknowledgement: “This work was supported by the National Cancer Institute of the National Institutes of Health under award number P30CA016086. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.”
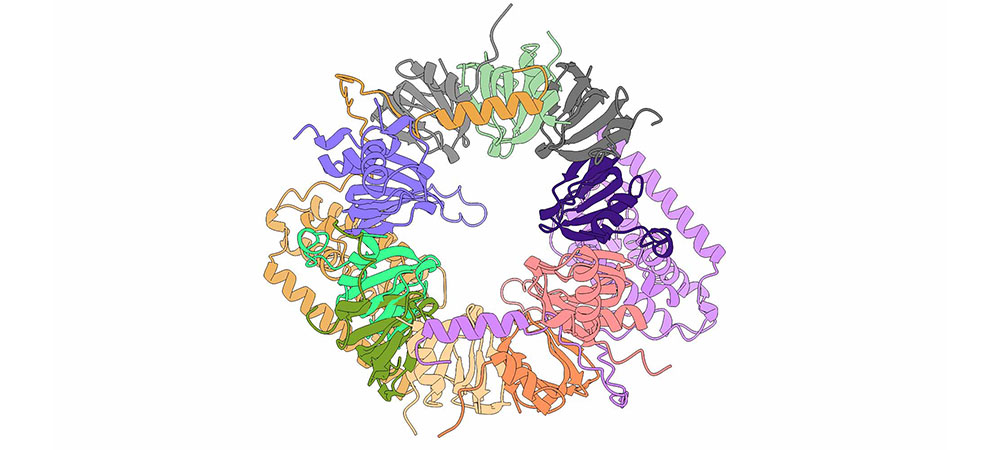
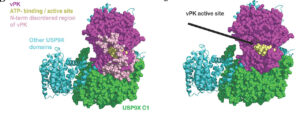
Image at the top of this page: This image depicts the SMN (Survival Motor Neuron) complex, a multi-protein assembly crucial for the biogenesis of small nuclear ribonucleoproteins (snRNPs). The complex is composed of several protein subunits intricately arranged to facilitate the assembly and function of snRNPs, which are essential components of the spliceosome machinery involved in pre-mRNA splicing.