Vironomics Core
SUBMISSION/DROP-OFF DEADLINES FOR 2021:
- 10/18/2021: Last day to drop-off RAW MATERIAL for library prep, pooling and sequencing for possible data delivery before University closure for winter holiday.
- 11/01/2021: Last day to drop-off STUDY MADE LIBRARIES for pooling and sequencing for possible data delivery before University closure.
- 11/15/2021: Last day to drop-off STUDY MADE POOLS for possible data delivery before University closure.
- 12/17/20201 at 1pm: Last day to DROP-OFF ALL sample types. We are closing drop off early to ensure that we can have everything checked in and stored properly before UNC closure.
- 12/20/2020 – 12/30/2021: Vironomics Core is closed. No submissions will be approved during this time.
- 01/10/2021: Vironomics Core reopens for 2022. Submissions will be approved and drop offs will begin on a first come first serve basis.
The Vironomics core is a core facility tailoring our resources to fit your individual project.
The Vironomics Core is led by our Principal Investigator, Dr. Dirk Dittmer, and includes several research staff. Our facilities are located on the 1st floor of the Lineberger Cancer Center (wing parallel with West Dr). The main research focus of the Dittmer lab is understanding viral tumorigenesis, especially that associated with Kaposi’s Sarcoma Herpesvirus (KSHV).
The Vironomics core operates all assays under good clinical laboratory practices (GCLP), which means we can generate information to support clinical trials and to support FDA approval, but cannot generate information that is reported back to the patient, or be used in treatment decisions.
News
-
Runaway Kaposi Sarcoma-associated herpesvirus replication correlates with systemic IL-10 levels
An avant-garde study of the uncommon KSHV-associated inflammatory cytokine syndrome. This paper was newly published in the Virology Journal at the beginning of this month.
-
Core sequencing QC
Quality Control plots for sequencing runs done in house by Vironomics Core’s on the Ion Torrent S5
-
UNC Super Collaboration
An example of a recent custom single-cell sequencing project that we assisted with. It included a large number of UNC collaborations including: Dr. De Paris lab, UNC Flow Cytometry Core, UNC Vironomics Core, and UNC HTSF. At this year’s Research Triangle Cytometry Association Meeting in January, Department of Microbiology and Immunology’s Dr. Kristina De Paris … Read more
-

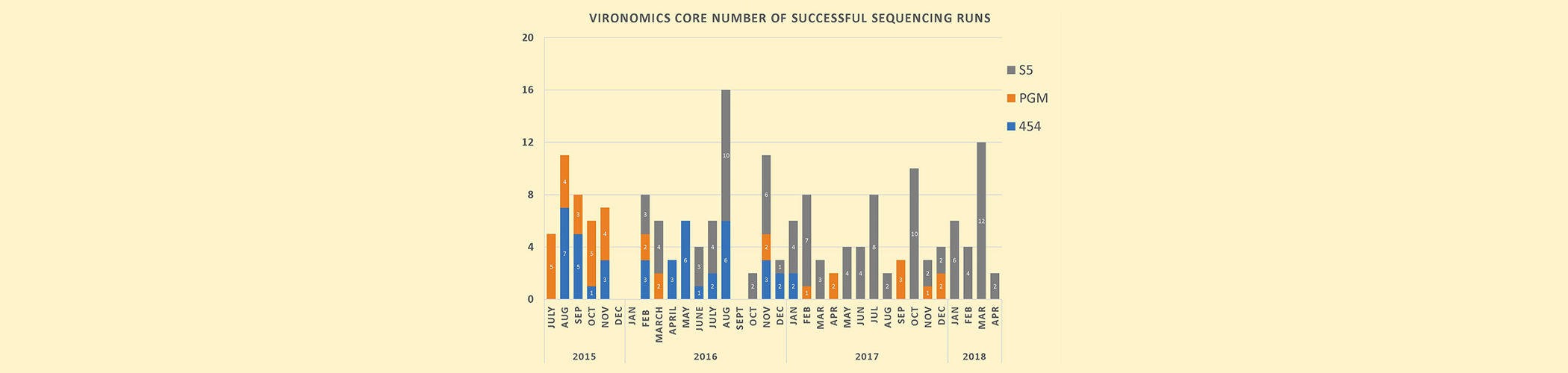
Plot of the successful sequencing runs
From July 2015-Feb 2017, the number of successful sequencing runs we’ve done in-house, generally with 1-2 staff.
Assays & Services
qPCR
The Vironomics Core currently provides several real-time qPCR arrays for viruses and miRNAs profiling including: KSHV, EBV, HSV1 and HSV2, RRV, HCMV, NFkappaB, Pre-miRNA, and P53response12. The unique automated robotics also enables them to offer custom and commercial real-time PCR assays to clients. The Vironomics Core uses several automated robotics for the large-scale real-time PCR. A Tecan Freedom Evo© is used to set up reactions in a 384-well plate, which is then run by the Roche Lightcycler 480 II©. This is a fast and effective way to amplify samples with RT-qPCR.
UNC Super Collaboration sequencing story
Sequencing
The Vironomics Core facilitates research long-read length next generation sequencing. The sequencers in the core are ideal for longer DNA fragment sequencing with instrument run times of only 24 hours. This is accomplished using Ion GeneStudio S5 Prime System, and companion Ion Torrent Chef. The Ion GeneStudio S5 capable of up to 600bp read lengths and has a maximal output of 130 Million 200 bp reads in 12 hours or 4 human exomes per day. Many clients use the longer reads for large genomes, de novo sequencing, 16S sequencing, long amplicons, etc. Customization is available or take advantage of the current expertise in: whole genome sequencing (WGS), de novo assembly, STR cell line verification, targeted amplicon sequencing, plasmid verification, strand-specific RNAseq, and exome sequencing.