Alternative Technologies
HTG Edge

At this time UNC HTSF only offers the sequencing for larger HTG library sets. We do have a collaborating core at NCCU that currently offers this service. Please contact Dr. Kumar at NCCU for library prep.
Please contact:
Lesli Colon — lrioscolon@nccu.edu
Elena Arthur — earthur1@nccu.edu
HTG Molecular’s EdgeSeq is a fully automated sample and library preparation platform for targeted RNA sequencing that pairs HTG’s extraction-free, high-specificity Edge Chemistry with the high sensitivity and dynamic range of next-gen sequencing. EdgeSeq enables digital quantitation of miRNA and mRNA expression from difficult sample types with libraries made directly from samples with out doing RNA isolation.
- Total RNA
- PAXgene RNA blood tubes
- formalin-fixed, paraffin-embedded (FFPE) tissues
- Fresh Frozen Tissue
- Plasma
- Serum
- Exosomes
- Cell Lines
- Buffy Coats
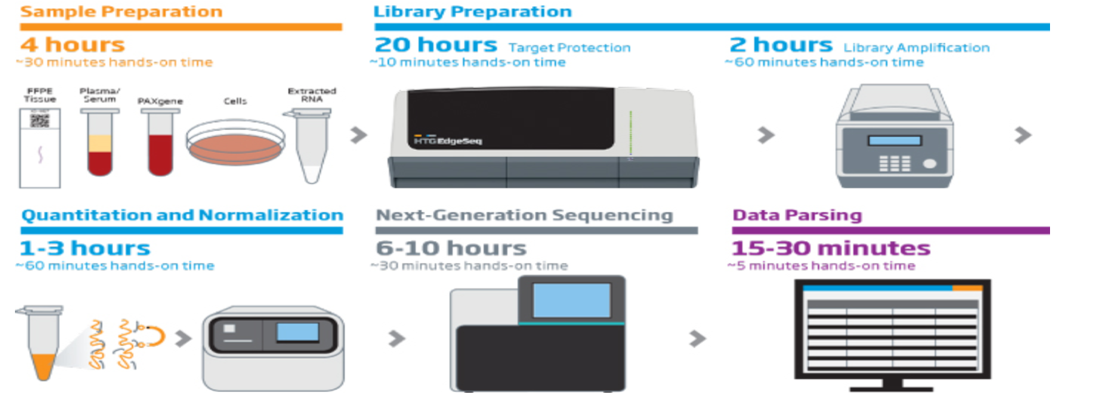
HTG‘s extraction-free chemistry reduces sample input requirements and preserves valuable specimens by eliminating timely and technically challenging steps such as cDNA synthesis, size selection, rRNA depletion, end repair, and adapter ligation.
HTG also offers two CE-IVD assays along with numerous RUO (research use only) assays. Listed below are the panels HTSF offer.
- HTG EdgeSeq Oncology Biomarker Panel (OBP)
- HTG EdgeSeq Precision Immuno-Oncology Assay (IO)
- HTG EdgeSeq miRNA Whole Transcriptome Assay (miRNA)
- HTG EdgeSeq PATH Panel (PATH)
- HTG EdgeSeq Lymphoma Panel (DLBCL)
- HTG EdgeSeq Mouse mRNA Tumor Response Panel (TRP)
Sequencing and pooling recommendations can be found in the chart below:
| Service | Samples per Pool | Platform | Cycles |
| HTG (2 million clusters per sample) | 8 | MiSeq | SE 50 |
| HTG (5 million clusters per sample) | 48 | HiSeq 2500 Rapid Run (full flowcell) | SE 50 |
| HTG (2, 5 million clusters per sample) | 96 | HiSeq 2500 Rapid Run (full flowcell) | SE 50 |
10X Genomics Chromium
The HTSF is excited to now offer a growing range of genomics applications for single cells, including RNA-seq, gene expression profiling by qPCR and DNA amplification for whole-genome or targeted (exome or PCR-based analysis) through 10x Genomics Chromium platform (similar to Drop Seq).
Sequencing technology as presented through the 10x Genomics Chromium Platform offers a high throughput molecular barcoding and analysis suite that delivers cell-by-cell 3’end counting of mRNA transcripts for many tens of thousands of cells per run- similar to the popular DropSeq technology. This technology supports a broad range of applications, including cancer-cell transcriptomics and cell-type identification and discovery. Because the platform works with short read sequencers, it integrates easily into the existing HTSF sequencing workflow allowing for an end to end service.
10x Technology overview:
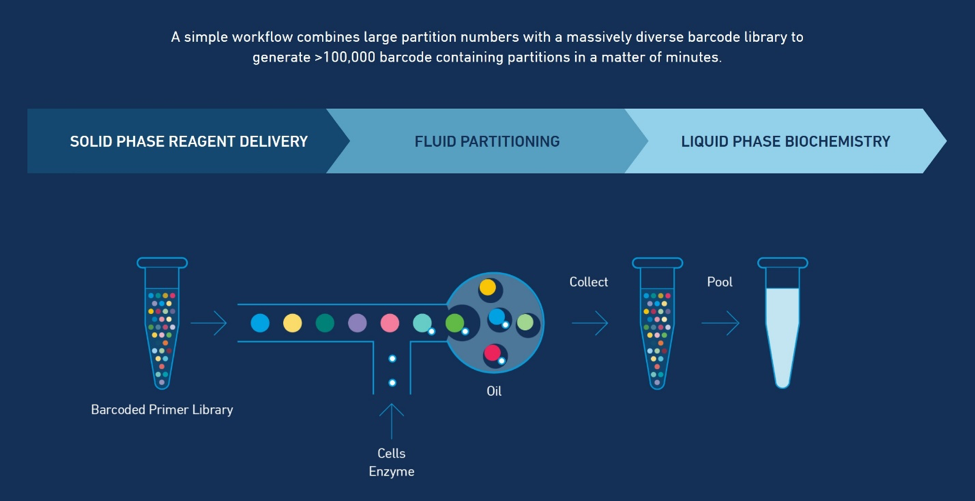
The 10x Genomics Chromium System, powered by GemCode Technology, provides a precisely engineered reagent delivery method that enables thousands of micro-reactions in parallel. Samples are encapsulated into hundreds to tens of thousands of uniquely addressable partitions each containing an identifying barcode for downstream analysis.
Each Gel Bead, infused with millions of unique oligonucleotide sequences, is mixed with a sample, which can be high molecular weight (HMW) DNA, individual cells, cells labeled using Feature Barcoding technology, nuclei, nuclei treated with transposase, or Cell Beads. Gel Beads and samples are then added to an oil-surfactant solution to create Gel Beads-in-emulsion (GEMs), which act as individual reaction vesicles in which the Gel Beads are dissolved and the sample is barcoded.
Barcoded products are pooled for downstream reactions to create short-read sequencer compatible libraries. After sequencing, the resulting barcoded short read sequences are fed into turnkey analysis pipelines that use the identifying barcodes to map reads back to their original HMW DNA, single cell, or single nucleus of origin.
Also see the overview – 10x BR025 Chromium Brochure or visit https://www.10xgenomics.com/products/
HTSF is not currently offering 10x Genomics protocols. HTSF does offer sequencing services for larger scale 10x Genomics projects. We accept study-made 10x libraries and those made by AACORE for other labs.
Please note that 10X whole genome has been discontinued.
Single Cell Gene Expression -The Chromium Single Cell Gene Expression Solution provides a comprehensive, scalable solution for cell characterization and gene expression profiling of hundreds to tens of thousands of cells. Dual indexes are now available for Chromium Single Cell Gene Expression libraries. Please see our 10x Library Indexes and Sequencing Requirements below for more information. It can help to uncover gene expression variability and identify rare cell types in heterogeneous samples to better characterize cellular contributions during development and disease. In the addition of barcoding, it is possible to:

- CRISPR Screening – identify cell-specific CRISPR-mediated perturbations on gene expression via direct capture of gRNAs and polyadenylated mRNAs from the same single cell
- Cell Surface Protein – measure both gene and cell surface protein expression in the same cell to identify protein isoforms, detect protein for low abundance transcripts, and further increase phenotypic specificity
Single Cell ATAC -Single Cell ATAC accelerates the understanding of the regulatory landscape of the genome, thereby providing insights into cell variability. The chromatin profiling of tens of thousands of single cells in parallel allows researchers to see how chromatin compaction and DNA-binding proteins regulate gene expression at high resolution.
Single Cell Multiome ATAC + Gene Expression -With the new Chromium Single Cell Multiome ATAC + Gene Expression, you can simultaneously profile gene expression and open chromatin from the same cell, across thousands of cells. Deepen your characterization of cell types and states with linked transcriptional and epigenomic analyses, discover new gene regulatory interactions and easily interpret epigenetic profiles with key expression markers.