Long Read Technologies
Oxford Nanopore
 At the High Throughput Sequencing Center, our goal is to provide the research community at UNC with access to cutting edge sequencing platforms. To that end, we are offering Oxford Nanopore Sequencing on the GridION platform.
At the High Throughput Sequencing Center, our goal is to provide the research community at UNC with access to cutting edge sequencing platforms. To that end, we are offering Oxford Nanopore Sequencing on the GridION platform.
Oxford Nanopore Technologies offers direct sequencing of native DNA or RNA, or samples that have been amplified with PCR. Nanopore sequencing can read any length of DNA/RNA, from short to ultra-long. It is useful for applications such as De novo assembly, scaffolding and finishing, bridging repetitive regions, structural variation, SNVs and phasing, targeted sequencing, RNA analysis, metagenomics, and epigenetics.
- A single MinION flow cell can produce reads up to 2 Mb long, and provide up to 10 Gb of sequencing data
- The GridION can run up to 5 MinION flow cells at one time
- There is no fixed run time; a user can run any of the systems for a short or long period of time, as data is streamed in real time
The Oxford Nanopore’s unique direct molecule sequencing platform is based on protein nanopores set in a polymer membrane. Current is passed through the nanopore, and as DNA/RNA is passed through the pore, a disruption in current is detected.
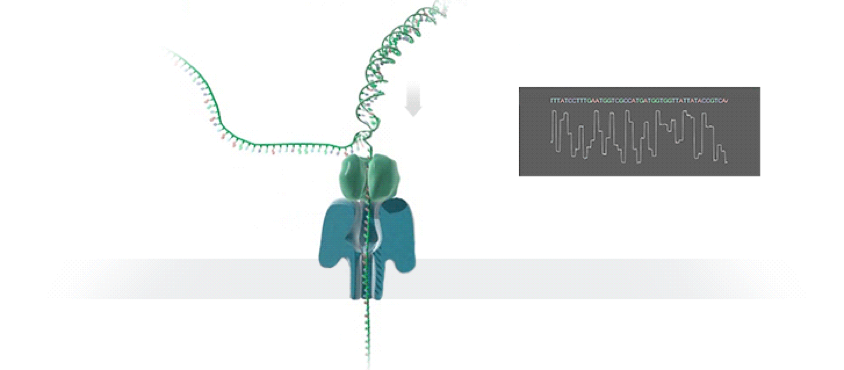
DNA Sequencing (from nanoporetech.com): A strand of DNA is passed through a nanopore. The current is changed as the bases G, A, T and C pass through the pore in different combinations.
Oxford Nanopore can produce ultra-long read sequencing reads, limited by DNA length/quality. In our hands we regularly generate reads >100kb. There are a host of applications, including:
- Rapid sequence identification
- Utilizing long reads for improved genome assembly
- Analysis of full length RNA transcripts from cDNA (PCR and PCR-free)
- Direct sequencing of RNA molecules
- Metagenomic analysis
- Structural variant detection
- Copy Number detection in complex regions
To learn more about Oxford Nanopore sequencing at the HTSF, read our white paper or contact Piotr Mieczkowski at Piotr_Mieczkowski@med.unc.edu or Tara Skelly at tskelly@email.unc.edu.
Bionano Genomics

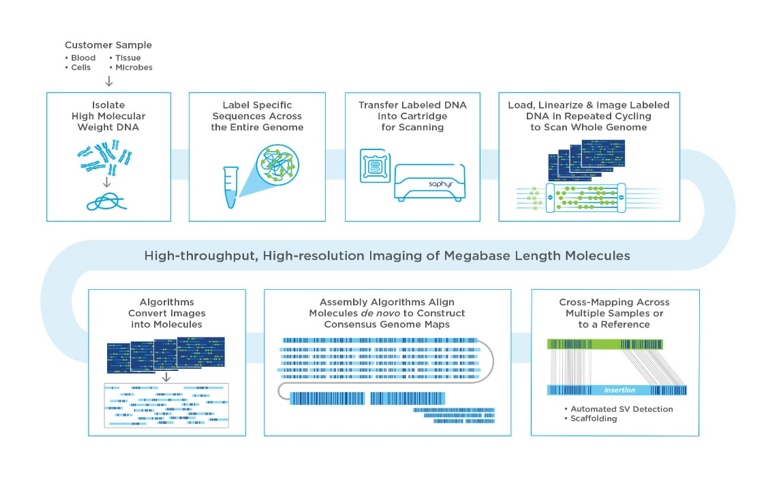
The HTSF also offers optical mapping with the introduction of the Bionano Saphyr instrument. Optical mapping can detect large-scale structural variations ranging from 500 bp to megabase pair lengths. Structural variations are responsible for many diseases, including cancer and developmental disorders. One Saphyr flow cell can produce up to 1300 Gb of data.
Applications:
- Structure of the cancer genome
- Structural variation in cell lines
- Genomes de novo assembly
- Genetic diseases
- Cytogenomics
- Agriculture
| Key applications | Optical mapping of whole genomes for structural variations and de novo assembly |
| Platform | Saphyr |
| Flowcells processed | 1 chip with 2 flowcells |
| Depth | Up to 1300 Gbp/flowcell (1300 Gbp only recommended for mixed population cancer. For standard human sized genomes 600 Gbp is recommended) |
| Run time | Until it reaches data target (usually less than 24 h) or up to 36h |
| Lanes/flowcell | 1 |
PLEASE NOTE:
- This is optical mapping not sequencing (https://bionanogenomics.com/products/saphyr/).
- The DNA is treated with an enzyme (DLE-1) that labels certain DNA sequences in the genome with florescent label and then the DNA is stained.
- The system maps the length of the DNA molecule (100 000 bp to Mbps) and where the labels are located on it creating a pattern of labels.
- Combining the molecules allows space to create map of the whole genome of the sample.
- The reference is marked digitally where the labels are expected to be and allows it to track any rearrangements in the sample. It can be also used for scaffolding of sequencing data.
- To learn more about optical mapping at the HTSF, please contact Piotr Mieczkowski at Piotr_Mieczkowski@med.unc.edu or Ewa Malc at ewa_malc@med.unc.edu
PacBio Sequel
UNC has a collaboration with NCSU’s Genomic Sciences Laboratory (GSL) to use their PacBio Sequel at the same internal cost you would receive if you were sequencing on UNC’s campus.For more information, see the GSL website
The PacBio system utilizes Single Molecule, Real-Time (SMRT) technology, eliminating the need for amplification. The DNA submitted serves as a direct template for the sequencing reaction via the ligation of hairpin adapters that create a circular template. Real-time sequencing occurs continuously across this circular template, producing extra-long read lengths and high consensus accuracy. These advantages make the PacBio Sequel a powerful tool in accurately calling SNPs and sequencing through CG-rich areas.
For more information, see the PacBio website
The PacBio Sequel produces up to 30 kb sequence reads with up to 7X more data output than the RS II. The Sequel System is ideal for rapidly and cost-effectively generating high-quality whole genome de novo assemblies, and facilitating full-length transcriptome sequencing (IsoSeq) efforts. Typical data output per SMRT Cell is 5-8 Gb.
For project quotes, questions and consults
Contact Dr. David (Andy) Baltzegar, GSL Director
(919)-513-0738 (8a – 4:30p Mon – Fri)
dabaltze@ncsu.edu
For instructions on submitting projects to the GSL, please visit https://research.ncsu.edu/gsl/sanger-sequencing/how-to-submit-a-project/