Advanced Analytics Core
The CGIBD’s Advanced Analytics (AA) Core provides high-quality, quantitative solutions for studying proteins and nucleic acids at the bulk or single-cell level. Whether measuring gene expression or cytokines, the AA core’s favorable pricing and unique cost-sharing model allow investigators access to a wide variety of assays and an extensive probe library for a fraction of their list price.
NEW FOR 2023: Epithelial Organoids and 2D Monolayer “EpCulture” Services:
UNC has pioneered several intestinal and colonic epithelial organoidand 2D monolayer culture platforms in studying the gastrointestinal tract. These platforms (AKA “EpCultures”) are powerful physiologically relevant in vitro models, but they can be expensive to implement due to recombinant growth factor requirements, difficulty in obtaining primary tissues (especially from human donors), and technical challenges for many laboratories. The Advanced Analytics Core is now proud to offer a broad range of services that will make these unique culture methods more accessible to investigators. More information available here.
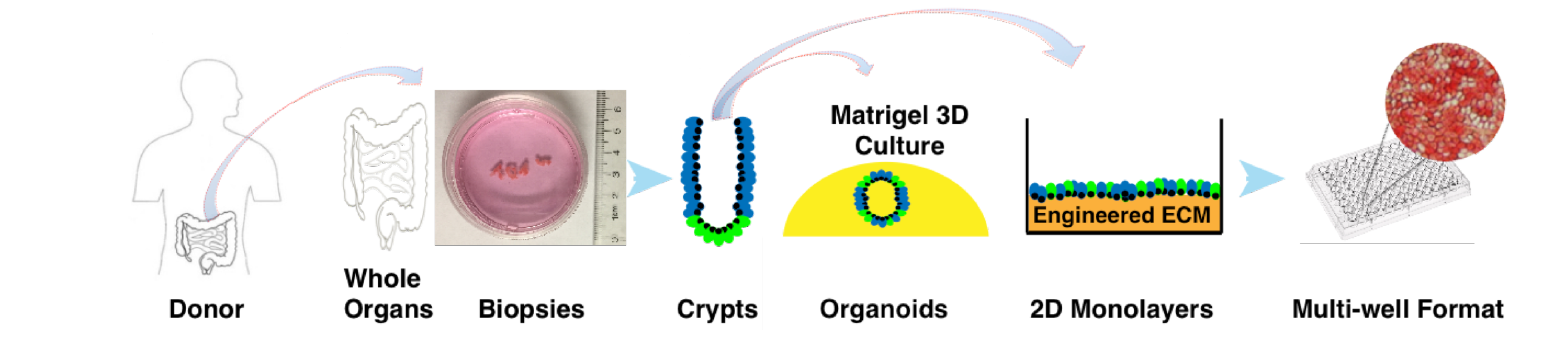
Organoid and 2D Monolayer “EpCulture” Services:
Conditioned Media (WRN and more)
Experimental Design Consultation
Bulk and single cell multi-omic services:
High-throughput PCR (Fluidigm/Standard BioTools BioMark HD)
–Gene Expression qPCR, SNP Genotyping, Copy Number Variation, Digital PCR
Library Preparation, Sequencing, and Basic Data Processing (10x Genomics Chromium, Fluidigm/Standard BioTools C1)
–RNA-Seq, DNA-Seq, ATAC-Seq, CITE-Seq, REAP-Seq
Protein Detection (Protein Simple Milo)
–Single Cell Western Blot
Cell sorting (FACS) – Available in conjunction with our other single cell services
Contacts:
Carlton Anderson, Associate Director
Research Specialist, FACS
Center for Gastrointestinal Biology and Disease
101 Mason Farm Rd, 030 Glaxo Bldg, CB# 7194
Chapel Hill, NC 27599
(919) 966-0175
Gabrielle Cannon
Research Specialist, Genomics (NGS and qPCR)
Center for Gastrointestinal Biology and Disease
111 Mason Farm Rd, 4331 MBRB, CB# 7032
Chapel Hill, NC, 27599
(919) 966-0175
Quinn Fong
Research Specialist, EpCultures
Center for Gastrointestinal Biology and Disease
111 Mason Farm Rd, 4331 MBRB, CB# 7032
Chapel Hill, NC, 27599
(919) 966-0175
Scott Magness, Director
Associate Professor
Departments of Medicine, Biomedical Engineering, and Cell & Molecular Physiology
Division of Gastroenterology and Hepatology
101A Mason Farm Rd, 4337 MBRB, CB# 7032
Chapel Hill, NC, 27599
(919) 966-6816
www.magnesslab.org