Johnson results
Gary Johnson
Research Synopsis:
My laboratory defines the “signal relay” systems initiated by various cellular stimuli including cytokines, growth factors, antigens, and drugs used to treat human disease. The unifying hypothesis directing the work in my laboratory is that the differential expression and spatial organization of signal relay proteins controls the responsiveness of different cell types to specific stimuli. Aberrant spatial and temporal control of signal relay systems is a major contributing factor to human diseases including cancer, cardiovascular disease and auto-immunity. The primary signal relay systems studied in the lab involve the mitogen activated protein kinase (MAPK) pathways and how they regulate gene expression, cell growth, development and apoptosis.
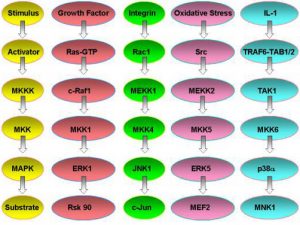
Fig.1. MAPK signal relay systems in mammalian cells.
MAP kinase pathways are composed of three kinases: a MAP kinase (MAPK), MAPK kinase (MKK), and a MAPK kinase kinase (MKKK). Activation of the MKKK by GTP binding regulatory proteins (i.e., Ras, Rac, Cdc42, Rho) or phosphorylation by another kinase (MKKKK, i.e., Mst1, PAK) controls the activity of a specific MAPK pathway. Currently, biochemical and genetic analysis has defined five different MAPK pathways in mammalian cells. The best understood include the extracellular-signal response kinase 1 and 2 (ERK1/2), ERK5, c-Jun NH2-terminal kinase (JNK) and p38 MAPK pathways. To date, 12 MAPKs, 7 MKKs, and 20 MKKKs have been identified in mammalian cells. Many of the MAPKs and MKKs are closely related kinases within the ERK, JNK, and p38 pathways. The MKKKs are more diverse allowing a differential control of specific MKK-MAPK component members to various cellular stimuli including cytokines, stress (i.e., heat shock, osmotic imbalance, hypoxia, drug exposure, etc.), and cell-cell contact. MKKKs also appear to be differentially expressed in different cell types; however, no detailed analysis of their expression has been published. Similarly, specific MKKs and MAPKs appear to be differentially expressed in cells of different tissue origin. The differential expression of the MKKKs, in particular, but also MKKs and MAPKs will dictate functional responses in different cell types to specific stimuli. The differential expression of MKKKs, for example, can allow one cell to respond with activation of the JNK pathway while another cell does not. To define the requirement and function of specific MAPK pathways in different cell types and in response to specific stimuli, we have used targeted gene disruption by homolgous recombination to abolish the expression of different MKKKs. These include MEKK1, 2, 3, and 4 which were originally cloned in my laboratory and selectively regulate the ERK1/2, ERK5, c-Jun kinase (JNK) or p38 pathways. More recently, we have made gene knockins to inactivate the kinase activity of specific MEKKs but the kinase-inactive protein is still expressed. In parallel with the gene knockout studies in mice, the laboratory uses live cell fluorescence imaging and fluorescence resonance energy transfer (FRET) to study the spatial and temporal activation, localization and function of MAPK pathways in cells. Combined with biochemical analysis of the proteins in MAPK signal relay systems, our studies define the role of MAPK pathways in the integrated response of cells to different stimuli and their physiological function in tissues and organs.
Recent Results:
An example of the power of gene knockouts to define the function of MAPK pathways in cellular regulation is the study of MEKK1. Only by using the knockout of MEKK1 were we able to define the role of MEKK1 in cell migration. MEKK1 is associated with actin fibers and focal adhesions, localizing MEKK1 to sites critical in the control of cell adhesion and migration. EGF-induced ERK1/2 activation and chemotaxis are inhibited in MEKK1-/- fibroblasts. MEKK1 deficiency causes loss of vinculin in focal adhesions of migrating cells, increased cell adhesion and impeded rear-end detachment. MEKK1 is required for activation of the cysteine protease calpain and cleavage of spectrin and talin, proteins linking focal adhesions to the cytoskeleton. Inhibition of ERK1/2 or calpain, but not JNK, mimics MEKK1 deficiency. Thus, the targeted disruption of MEKK1 expression directly demonstrated that MEKK1 regulates calpain-mediated substratum release of migrating cells.

|
Fig. 1. Defective cell migration in cells deficient in MEKK1 protein expression. Wild type or MEKK1-/- fibroblasts were seeded on glass coverslips and grown to confluency. In addition, add-back fibroblasts having the full length MEKK1 protein re-expressed in the MEKK1-/- fibroblasts were also allowed to grow to confluency. A wound in the culture was generated with the swipe of a razor blade. Wild type and MEKK1 add-back fibroblasts readily migrate into the wound space. MEKK1-/- fibroblasts are unable to migrate into the wound space demonstrating their migration is inhibited. |
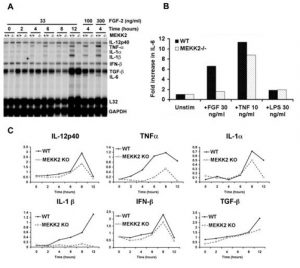
In contrast to the function of MEKK1 in the control of cell migration, the targeted disruption of MEKK2 demonstrated a different function for this MKKK. MEKK1 coordinately regulates ERK1/2 and JNK. In contrast, we discovered that MEKK2 coordinately activates the ERK5 and JNK pathways. Targeted disruption of MEKK2 expression causes loss of ERK5 and JNK activation in response to FGF-2 in mouse embryonic fibroblasts. FGF-2 receptor signaling requires MEKK2 for induction of mRNA for c-Jun, Fra-1, and Fra-2, components of the AP-1 transcription complex. In FGF-2-stimulated MEKK2-/- fibroblasts, c-Jun phosphorylation is inhibited, consistent with a loss of JNK activation. Thus, MEKK2 regulates AP-1 activity at two levels, by regulating both expression of AP-1 components and c-Jun N-terminal phosphorylation. One function of the AP-1 transcription complex is to regulate cytokine gene expression. Expression of IL-1 a , IL-1 b , IL-6 and TNF a is inhibited in MEKK2-/- fibroblasts. Bacterial lipopolysaccharide (LPS) and TNF a neither activate ERK5 nor require MEKK2 for JNK activation, demonstrating specificity of MEKK2 in FGF-2 receptor signaling and control of cytokine gene expression. Similar gene knockout studies with MEKK3 and MEKK4 have shown their selective regulation of MAPK pathways for the control of additional regulatory responses in different cell types. This is an ongoing area of research in the laboratory.
|
Fig. 2. FGF-2 regulation of cytokine expression in WT and MEKK2-/- MEFs. A. Cytokine mRNA expression was analyzed using an RPA probe set for IL-12p40, TNF- a , IL-1 a , IL-1 b , interferon- b (IFN- b ), TGF- b , and IL-6, with L32 and GAPDH as normalization controls. Results show a substantial reduction in specific cytokine mRNA expression and in response to FGF-2 for TNF- a , IL-1 a , IL-1 b , and IL-6 in the MEKK2-/- MEFs. IL-12, IFN- b , and TGF- b mRNA levels were not affected by loss of MEKK2 expression. B. ELISA for IL-6 expression in the culture supernatants 24 hours following stimulation with FGF-2, TNF a, or LPS. Results show a significant decrease in IL-6 production in the MEKK2-/- compared with wild type MEFs. Stimulation with TNF- a resulted in the production of IL-6 protein at similar levels for MEKK2-/- and wild type MEFs. LPS did not stimulate the production of IL-6 protein in either wild type or MEKK2-/- MEFs. C. Quantitation of the RPA analysis shown in (A) by phosphorimaging, with normalization for L32 and GAPDH mRNA expression levels. The results are representative of three independent experiments. |
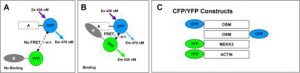
Another research strategy in the lab is to define the spatial localization and regulation of signal relay systems in cells. One approach to do this is the use of live cell imaging using proteins tagged with CFP and YFP. The value of the mutant green fluorescent proteins, YFP and CFP, is that they can be imaged separately in the same cell because of their different excitation and emission spectra. Importantly, under tight distance constraints, energy can be transferred from excited CFP donor to YFP acceptor through a non-radiative dipole-dipole interaction (FRET). There is a strong inverse relationship between FRET and chromophore separation distance such that CFP-YFP FRET (CYFRET) occurs only if the two proteins are brought into close proximity (
|
Fig. 3. FRET measures protein interactions in cells. CYFRET technology: CYFRET is a powerful technique to measure the interaction of two proteins in both live cells and paraformaldehyde fixed cells. CYFRET between two proteins occurs only when the two proteins are in close proximity in a range of 50 Å or less. |
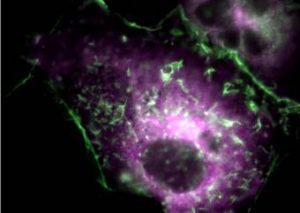
We have discovered a novel scaffold protein that binds actin, the GTPase Rac and the upstream kinases MEKK3 and MKK3 in the p38 MAPK phosphorelay module. RNAi demonstrated that MEKK3 and the scaffold protein are required for p38 activation in response to sorbitol-induced hyperosmolarity. We used FRET to show a cytoplasmic complex of the MEKK3-scaffold protein that is recruited to dynamic actin structures in response to sorbitol. By virtue of its ability to bind actin, relocalize to Rac-containing membrane ruffles and its obligate requirement for p38 activation in response to sorbitol, we termed this protein O smosensing S caffold for M EKK3 (OSM). The Rac-OSM-MEKK3-MKK3 complex is the mammalian counterpart of the CDC42-STE50-STE11-Pbs2 complex in Saccharomyces cerevisiae for the regulation of p38 activity.
|
Fig. 4. Recruitment of a p38 MAPK signaling scaffold (OSM) to actin filaments at sites of membrane ruffling in response to hyperosmolar stress. Actin is represented as YFP-actin and OSM is CFP-OSM. OSM organizes a MAPKKK (MEKK3), a MKK (MKK3) and the GTPase, Rac1 for the localized activation of p38 kinase activity. |
Recent Publications:
Witowsky, J.A., Johnson, G.L. (2003) Ubiquitylation of MEKK1 inhibits its phosphorylation of MKK1 and MKK4 and activation of the ERK1/2 and JNK pathways. J. Biol. Chem . 278, 1403-1406.
Witowsky, J.A., Abell, A., Johnson, N.L., Johnson, G.L. , Cuevas, B.D. (2003) MEKK1 is required for inducible urokinase-type plasminogen activator expression. J. Biol. Chem . 278, 5941-5946.
Sun, W., Wei, X., Kesavan, K., Garrington, T.P., Fan, R., Mei, J., Anderson, S.M., Gelfand, E.W., Johnson, G.L. (2003) MEK kinase 2 and the adaptor protein Lad regulate extracellular signal-regulated kinase 5 activation by epidermal growth factor via Src. Molecular and Cell Biol. 23, 2298-22308.
Cuevas, B.D., Abell, A.N., Witowsky, J.A., Yujiri, T., Johnson, N. L., Kesavan, K., Ware, M., Jones, P.L., Weed, S.A., DeBiasi, R. L., Oka, Y., Tyler, K.L. and Johnson, G.L. (2003) MEKK1 regulates calpain-dependent proteolysis of focal adhesion proteins for rear-end detachment of migrating fibroblasts. EMBO J. 22, 3346-3355.
DeBiasi, R.L., Clarke, P., Meintzer, S., Jotte, R., Kleinschmidt-Demasters, B.K., Johnson, G.L. and Tyler, K.L. (2003) Reovirus-induced alteration in expression of apoptosis and DNA repair genes with potential roles in viral pathogenesis. J. Virol ., 77, 8934-8947.
Nakamura, K. and Johnson, G.L. (2003) PB1 domains of MEKK2 and MEKK3 interact with the MEK5 PB1 domain for activation of the ERK5 pathway. J. Biol. Chem. 278, 36989-36992.
Wei, X., Sun, W., Fan, R., Hahn, J., Joetham, A., Li, G., Webb, S., Garrington, T., Dakhama, A., Lucas, J., Johnson, G.L. and Gelfand, E.W. (2003) MEF2C regulates c-Jun but not TNF- a gene expression in stimulated mast cells. Eur. J. Immunol. 33, 2903-2909.
Abell, A.M., Knall, C., Ambruso, D.R., Riches, D.W. and Johnson, G. L. (2003) Rac2 D57N , a dominant negative mutant that inhibits signaling and disrupts the actin cytoskeleton. J. Cell Science , in press.
Kamala , K., Lobel-Rice, K., Sun, W., Lapadat, R., Webb, S., Johnson, G.L. and Garrington, T.P. (2003) MEKK2 regulates the coordinate activation of ERK5 and JNK in response to FGF-2 in fibroblasts. J. Cell. Physiol. , in press.
Uhlik, M.T., Abell, A.N., Johnson, N.L., Sun W., Cuevas, B.D., Lobel-Rice,K.E., Horne, E.A., Dell’Acqua, M.L. and Johnson, G.L. (2003) Rac-MEKK3-MKK3 Scaffolding for p38 MAPK Activation During Hyperosmotic Shock. Nat. Cell Biol. 5, 1104-1110.
