Research Interests
Key words: mouse models of human disease, chromatin-modifying factors, epigenetics
1. Investigating the Role of SWI/SNF Chromatin-Remodeling Complexes in Mammalian Development and Disease
Sequence-specific transcription factors are necessary, but usually not sufficient, to regulate transcription because the RNA Polymerase II (Pol II) holoenzyme cannot access promoter DNA in the context of nucleosomes or higher-order chromatin structures. However, transcriptional activators bound to enhancers or promoters can recruit SWI/SNF chromatin-remodeling complexes, and the BRG1 catalytic subunit utilizes its DNA-dependent ATPase activity to alter the conformation and position of nucleosomes. As a result, these promoters adopt an “open” chromatin configuration capable of binding Pol II so transcription can be initiated. Conversely, transcriptional repressors can recruit similar or identical BRG1-containing SWI/SNF complexes to other loci to establish a “closed” chromatin configuration that precludes transcription.
SWI/SNF complexes have been studied by analyzing how recombinant BRG1 protein and other subunits (called BRG1 associated factors or BAFs) affect artificial nucleosome arrays in cell-free systems. Although these studies have provided valuable information about the mechanism of nucleosome remodeling, they have not provided insight into the function of SWI/SNF complexes in vivo. Our primary objective is to understand the role of SWI/SNF complexes in regulation of gene expression during mammalian development and disease. The approach we are taking is to analyze an allelic series of Brg1 mutations in mice. We created a null allele by gene targeting and demonstrated that homozygotes die at the peri-implantation stage and heterozygotes are susceptible to exencephaly and mammary gland tumors. To circumvent the early embryonic lethality of null homozygotes, we have generated an ENU-induced hypomorph allele and several Cre/loxP conditional mutant lines of mice. We are particularly interested in hematopoiesis. The availability of powerful tools and reagents (e.g., flow cytometery and cell-surface markers, bone marrow transplantation of irradiated or immune-deficient hosts) has made the hematopoietic system a valuable model for studying factors involved in the proliferation of stem cells and their differentiation into various lineages in vivo. Some of the current projects being developed around our Brg1 mutations are outlined below:
a. The mechanism of BRG1 nucleosome remodeling:
We performed a whole-animal ENU mutagenesis screen and isolated a Brg1 point mutation (Brg1ENU1) that results in a single amino-acid substitution in the ATPase domain (E1083G). The mutant BRG1 protein is stable, assembles into SWI/SNF complexes, and exhibits normal ATPase activity but is unable to establish DNaseI hypersensitive sites characteristic of open chromatin. In other words, this mutation uncouples ATPase activity from nucleosome remodeling. The E1083G substitution likely interferes with the ability of BRG1 to harness the energy of ATP hydrolysis to remodel nucleosomes. To understand the molecular nature of BRG1 function, we will compare wild-type and mutant BRG1 protein in nucleosome remodeling assays.
b. Regulation of globin gene expression and erythropoiesis:
Brg1null/ENU1 mutants have a severe defect in fetal-liver erythropoiesis and die at embryonic day (E) 12.5. Mutant BRG1 complexes are recruited to the beta-globin locus control region (LCR), but the LCR does not adopt an open chromatin conformation while histone acetylation is decreased and DNA methylation is increased. Consistent with these aberrant epigenetic modifications, beta-globin transcription is severely reduced to ~5% of normal. ChIP assays will be performed to determine whether this mutation affects the occupancy of several erythroid transcription factors and chromatin-modifying factors at specific sites in the LCR and beta-globin promoters. The aim of this work is to understand the role of BRG1 in the multi-step beta-globin activation mechanism.
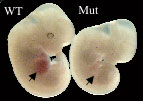 Wild-type (WT) and mutant (Mut) E12.5 embryos. Hemoglobin-containing erythroid cells are abundant in the WT fetal liver (arrowhead) but absent in the Mut fetal liver (arrowhead). In mutants, there is a block in differentiation at the basophilic erythroblast to polychromatic erythroblast transition, which coincides with hemoglobin production.
Wild-type (WT) and mutant (Mut) E12.5 embryos. Hemoglobin-containing erythroid cells are abundant in the WT fetal liver (arrowhead) but absent in the Mut fetal liver (arrowhead). In mutants, there is a block in differentiation at the basophilic erythroblast to polychromatic erythroblast transition, which coincides with hemoglobin production.
Beta-globin transcription is also reduced in the erythroid lineage of Brg1null/ENU1 mutants so we plan to evaluate the chromatin structure of the beta-globin major regulatory element (MRE). Finally, Brg1ENU1/ENU1 homozygotes come to term but are runted, fail to thrive, and often die before weaning. We plan to determine whether an erythropoiesis defect or something else is responsible for this postnatal phenotype.
c. Regulation of cytokine expression and chronic inflammation:
We have been studying a Brg1 conditional mutant line (Brg1MMTV-Cre) that exhibits severe skin inflammation. Brg1 is inactivated in the skin/hair follicle as well as the immune system of these mice. Their skin and hair follicles are hyperkeratinized based on analysis of H&E-stained skin sections and immunohistochemistry. Consequently, hair follicles break, get trapped between the epidermis and dermis, and apparently trigger an autoimmune response. Although there are a normal number of Treg cells, their function may be compromised and there is an increased number of monocytes/macrophages, granulocytes, and activated T cells. Preliminary evidence suggests that IL10 is downregulated. To understand the molecular basis of this inflammatory phenotype, we will identify BRG1 binding sites in skin and cells of the immune system using ChIP-on-chip. We will also focus on downstream candidate genes that have already been identified such as cytokeratins and IL10. Finally, we will perform bone marrow transplantation experiments or inactivate Brg1 only in the skin or the immune system (using transgenic or knock-in mouse lines that express Cre in a restricted manner) to address the relative importance of these two tissues in the inflammatory response.
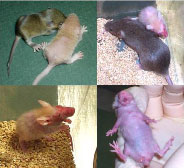 The MMTV-Cre transgene is linked to a coat-color locus so mutants are beige instead of agouti. The mutant skin has a flaky appearance and the hair does not come in properly, particularly near the hindquarters (upper left). Mutants are often runted compared to control siblings after weaning (upper right). Mutants have patchy hair growth and their skin is inflamed and ulcerated, particularly around the face, throat, and upper chest region (lower left and right).
The MMTV-Cre transgene is linked to a coat-color locus so mutants are beige instead of agouti. The mutant skin has a flaky appearance and the hair does not come in properly, particularly near the hindquarters (upper left). Mutants are often runted compared to control siblings after weaning (upper right). Mutants have patchy hair growth and their skin is inflamed and ulcerated, particularly around the face, throat, and upper chest region (lower left and right).
d. Cancer:
Brg1 null heterozygotes are susceptible to mammary tumors that exhibit genomic instability. Our molecular analyses indicate that Brg1 is a haploinsufficient tumor suppressor rather than undergoing loss of heterozygosity (LOH). To increase the penetrance and/or reduce the latency period, we are crossing this mutation to other mouse models of breast cancer. We are also analyzing the Wap-Cre conditional mutant line where both Brg1 alleles are inactivated in mammary tissue.
2. Investigating the Role of Dietary Fiber, Gut Microflora, and Epigenetic Events that Protect Against Colorectal Cancer
It is controversial whether dietary fiber protects against colorectal cancer (CRC) because of conflicting evidence from human epidemiological studies. Genetic heterogeneity and differences in gut microflora among participants have confounded these studies. To complicate matters further, these studies have utilized different sources of dietary fiber (which are fermented by gut microflora into different metabolites) and have not considered other dietary factors that might mask a beneficial fiber effect such as total caloric intake or dietary fat. To determine unequivocally whether dietary fiber protects against CRC, we are using several mouse models (ApcMin/+, AOM, and AOM/DSS) because their genetics, gut microflora, and diet can be rigorously controlled. For each model, we will provide low- or high-fiber diets to inbred mice that have defined gut microflora or are maintained germfree at the UNC Gnotobiotic facility.
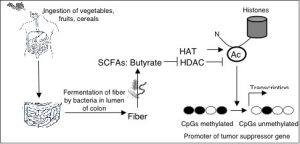 Because we are particularly interested in determining whether butyrate is an important metabolite, we will also colonize germfree mice with different bacterial strains that produce high or low levels of butyrate from fiber. These experiments will allow dietary fiber and gut microflora to be correlated with butyrate levels and tumor number and size. The different mouse models being analyzed will allow us to determine whether dietary fiber and gut microflora have a direct effect on the colonic epithelium and/or an indirect effect mediated by the immune system and inflammation.
Because we are particularly interested in determining whether butyrate is an important metabolite, we will also colonize germfree mice with different bacterial strains that produce high or low levels of butyrate from fiber. These experiments will allow dietary fiber and gut microflora to be correlated with butyrate levels and tumor number and size. The different mouse models being analyzed will allow us to determine whether dietary fiber and gut microflora have a direct effect on the colonic epithelium and/or an indirect effect mediated by the immune system and inflammation.
It is our hypothesis that dietary fiber has a protective effect because it is fermented into butyrate, which is a potent histone deacetylase (HDAC) inhibitor that affects gene expression. Therefore, we will identify fiber- and butyrate-induced changes in CpG methylation and gene expression that occur in normal and cancerous colon tissue from mice provided different diets and colonized with different gut microflora. CpG methylation will be analyzed in a genome-wide manner using MeDIP and tiling microarrays, while expression profiling will be performed using Agilent microarrays. Aberrant CpG methylation of many tumor suppressor genes constitutes the “second hit” to achieve loss of heterozygosity (LOH). Considering butyrate is an HDAC inhibitor, covalent histone modifications will be changed prior to CpG methylation. To understand the mechanism, we will perform ChIPs and other assays to determine the sequence of epigenetic events that occur at genes relevant to CRC in response to dietary fiber and gut microflora in normal and cancerous colon tissue.
Publications
Trainees
- Dallas Donahoe, Postdoctoral Fellow, Email
- Gary Rosson, Research Assistant, Email
- Darcey Wood Holley, Research Assistant, Email
Scott Bultman in UNC Genetics News

November 2, 2020
Department of Genetics Publications for October 11-31, 2020
Department of Genetics faculty, postdocs, students and collaborators published 18 papers during October 11-31, 2020. Survival, Pathologic Response, and Genomics in CALGB 40601 (Alliance), a Neoadjuvant Phase III Trial of Paclitaxel-Trastuzumab With or Without Lapatinib in HER2-Positive Breast Cancer. Fernandez-Martinez A, Krop IE, Hillman DW, Polley MY, Parker JS, Huebner L, Hoadley KA, …

August 10, 2020
Department of Genetics Publications for July 26 – August 8, 2020
Department of Genetics faculty, postdocs, students and staff published nine papers during July 26 – August 8, 2020.

June 15, 2020
Department of Genetics Publications for May 31 – June 13, 2020
Department of Genetics faculty, postdocs, students and collaborators published eleven papers during May 31 – June 13, 2020.

March 23, 2020
Department of Genetics Publications for March 8-21, 2020
Department of Genetics faculty, postdocs, students and collaborators published twelve papers during March 8-21, 2020.

July 29, 2019
Department of Genetics Publications for July 14-27, 2019
Department of Genetics faculty, postdocs, students and collaborators published ten papers during July 14-27, 2019.

