Professor of Biochemistry and Biophysics
(PhD – University of California, San Diego)
ACCEPTING STUDENTS
HONORS & AWARDS
- Fellow, American Association for the Advancement of Science (AAAS) 2013
- American Institute of Physics Board of Directors, 2012-present
ACTIVE PROJECTS
Carter lab has funding from two Foundations for projects arising from interest in the origin and evolution of the Genetic Code (Alfred P. Sloan Foundation, $1.5 M) and the emergence of codon-directed protein synthesis (W. M. Keck Foundation, $1 M) prior to the appearance of ribosomes.
RESEARCH
Protein Crystallography, Structural Polymorphism and Function
The Carter lab uses structural, bioinformatic, molecular genetic, and biophysical techniques to strengthen and deepen understanding of the mechanistic basis and historical origins of enzyme catalysis. See also Carter lab website.
Our structural biology research focuses on two problems:
How did the proteome emerge from pre-biotic chemistry?
Class I and II aminoacyl-tRNA synthetase superfamilies (aaRS). Translate the genetic code. Experimental properties of their deconstructed modules allow us to characterize development of catalysis from ~50 residue peptides. Multiple evidence argues that the two classes descended from opposite strands of a common ancestral gene. Unique information in that gene had two different, functional interpretations, depending on which strand is translated. The duality evokes visual puzzles ((Fig. 1; Martinez, et al. (2016); Carter, (2016)).
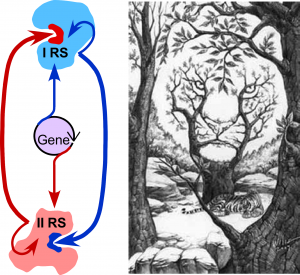
Figure 1. Dual genetic ancestry and catalytic interdependence of Class I and II aaRS.
Correlations between amino acid properties (Wolfenden), their exposed surface areas in folded proteins, and coding elements in transfer RNA clarify the “chemical ecology” of the Central Dogma (Fig. 2). Amino acid side chain behaviors determine folding of aaRS, giving them their reflexive ability to execute coding rules (curved arrow; Carter & Wolfenden (2016)).
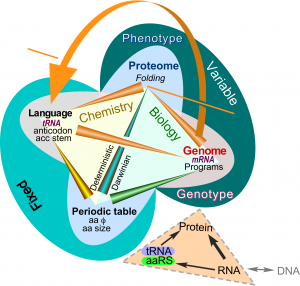
Figure 2. Crick’s Central Dogma and adaptor hypothesis in the ecology of amino acid phase transfer equilibria.
How do long range intramolecular communications produce vectorial behavior in enzymes that transduce chemical free energy?
Catalytic conformational changes during tryptophan activation by Tryptophanyl-tRNA synthetase (TrpRS) become favorable only after release of PPi (Fig. 3, b,d; with Dokholyan group). Axes are conformational angles—hinge bending and twisting—accounting for 0.85 of the motion. Red spheres denoting the conformational transition states found by PATH program simulations match the saddle point of the free-energy surfaces. Unless the PPi leaving group dissociates, the equilibrium favors the pre-transition state at high twist. (See Carter, et al. (2017) Chandrasekaran & Carter (2017).
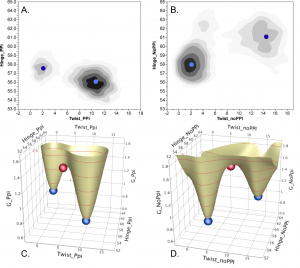
Figure 3. Conformational Free energy landscapes depend on PPi product release.
High-order combinatorial permutation measures long-range coupling free energies responsible for catalysis and specificity. Experimental ΔGkcat values correlate with structural perturbations in 15 combinatorial mutations at 4 sites that impose the barrier between pre-transition and products states, and with parameters from computational trajectories. The correlations relate catalysis to structure directly and via the computed trajectories (see Chandrasekaran and Carter (2017).
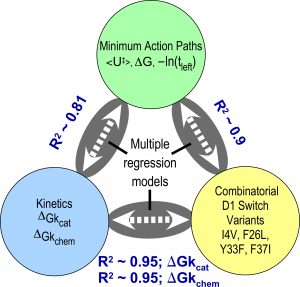
Figure 4. Structural changes correlate closely with experimental, computational parameters.
SELECTED PUBLICATIONS
(click for full publication list)
- A Zymography technique to study amino acid activation by aminoacyl-tRNA synthetases (aaRS): A broad spectrum, high-throughput tool to screen activities of aaRS and their Urzyme …
SK Patra, CW Carter Jr bioRxiv, 2023.02. 01.526722 - How did the Proteome Emerge From Pre-biotic Chemistry? CW Carter Jr Prebiotic Chemistry and Life’s Origin, 317-346 2022
- A Leucyl-tRNA Synthetase Urzyme: Authenticity of tRNA Synthetase Catalytic Activities and Promiscuous Phosphorylation of Leucyl-5′ AMP JJ Hobson, Z Li, H Hu, CW Carter Jr International Journal of Molecular Sciences 23 (8), 4229 2022
- Multidimensional Phylogenetic Metrics Identify Class I Aminoacyl-tRNA Synthetase Evolutionary Mosaicity and Inter-Modular Coupling CW Carter Jr, A Popinga, R Bouckaert, PR Wills International Journal of Molecular Sciences 23 (3), 1520 2022
- The roots of genetic coding in aminoacyl-tRNA synthetase duality CW Carter Jr, PR Wills Annual review of biochemistry 90, 349-373 2021
- Simultaneous codon usage, the origin of the proteome, and the emergence of de-novo proteins CW Carter Jr Current Opinion in Structural Biology 68, 142-148 2021
- Reciprocally-coupled gating: Strange loops in bioenergetics, genetics, and catalysis CW Carter Jr, PR Wills Biomolecules 11 (2), 265 2021
- Impedance matching and the choice between alternative pathways for the origin of genetic coding
PR Wills, CW Carter Jr International Journal of Molecular Sciences 21 (19), 7392 2020 - Escapement mechanisms: Efficient free energy transduction by reciprocally‐coupled gating CW Carter Jr Proteins: Structure, Function, and Bioinformatics 88 (5), 710-717 2020
- High-Resolution, Multidimensional Phylogenetic Metrics Identify Class I Aminoacyl-tRNA Synthetase Evolutionary Mosaicity and Inter-modular ‘Coupling CW Carter Jr, A Popinga, R Bouckaert, PR Wills BioRxiv, 2020.04. 09.033712
For Compilations of Carter lab publications, Selected publications by topic: visit Carter lab website

