GaMD-Amber: Manual
Download a PDF Manual of the latest GaMD, LiGaMD, Pep-GaMD and PPI-GaMD in Amber.
GaMD Algorithm
GaMD {
If (irest_gamd == 0) then
For i = 1, …, ntcmd // run initial conventional molecular dynamics
If (i >= ntcmdprep) Update Vmax, Vmin
If (i >= ntcmdprep && i%ntave ==0) Update Vavg, sigmaV
End
Save Vmax,Vmin,Vavg,sigmaV to “gamd_restart.dat” file
Calc_E_k0(iE,sigma0,Vmax,Vmin,Vavg,sigmaV)
For i = ntcmd+1, …, ntcmd+nteb // Run biasing molecular dynamics simulation steps
deltaV = 0.5*k0*(E-V)**2/(Vmax-Vmin)
V = V + deltaV
If (i >= ntcmd+ntebprep) Update Vmax, Vmin
If (i >= ntcmd+ntebprep && i%ntave ==0) Update Vavg, sigmaV
Calc_E_k0(iE,sigma0,Vmax,Vmin,Vavg,sigmaV)
End
Save Vmax,Vmin,Vavg,sigmaV to “gamd_restart.dat” file
else if (irest_gamd == 1) then
Read Vmax,Vmin,Vavg,sigmaV from “gamd_restart.dat” file
End if
For i = ntcmd+nteb+1, …, nstlim // run production simulation
deltaV = 0.5*k0*(E-V)**2/(Vmax-Vmin)
V = V + deltaV
End
}
Subroutine Calc_E_k0(iE,sigma0,Vmax,Vmin,Vavg,sigmaV) {
if iE = 1 :
E = Vmax
k0’ = (sigma0/sigmaV) * (Vmax-Vmin)/(Vmax-Vavg)
k0 = min(1.0, k0’)
else if iE = 2 :
k0” = (1-sigma0/sigmaV) * (Vmax-Vmin)/(Vavg-Vmin)
if 0 < k0” <= 1 :
k0 = k0”
E = Vmin + (Vmax-Vmin)/k0
else
E = Vmax
k0’ = (sigma0/sigmaV) * (Vmax-Vmin)/(Vmax-Vavg)
k0 = min(1.0, k0’)
end
end
}
LiGaMD Algorithm
LiGaMD {
If (irest_gamd == 0) then
For i
= 1, …, ntcmd // run initial conventional molecular
dynamics
If (i >= ntcmdprep) Update Vmax, Vmin
If (i >= ntcmdprep && i%ntave ==0) Update Vavg, sigmaV
End
Save Vmax,Vmin,Vavg,sigmaV
to “gamd_restart.dat” file
Calc_E_k0(iE,sigma0,Vmax,Vmin,Vavg,sigmaV)
For i = ntcmd+1, …, ntcmd+nteb // Run
biasing molecular dynamics simulation steps
deltaV = 0.5*k0*(E-V)**2/(Vmax-Vmin)
V = V
+ deltaV
If (i >= ntcmd+ntebprep) Update
Vmax, Vmin
If (i >= ntcmd+ntebprep &&
i%ntave ==0) Update Vavg, sigmaV
Calc_E_k0(iE,sigma0,Vmax,Vmin,Vavg,sigmaV)
End
Save Vmax,Vmin,Vavg,sigmaV
to “gamd_restart.dat” file
else if (irest_gamd == 1)
then
Read Vmax,Vmin,Vavg,
sigmaV from “gamd_restart.dat” file
End if
ntwin = ntmsd+nftau*ntwx
lig0=1 // ID of
the bound ligand
For i
= ntcmd+nteb+1, …, nstlim // run production
simulation
If (ibblig == 1
&& i%ntwx ==0) then // identify the bound
ligand according to the distance to protein
For ilig
= 1, …, nlig
dlig = distance(atom_p, atom_l)
If (dmin <= dlig) blig_min=ilig; dmin=dlig
End
If (dmin
<= dblig) blig=blig_min
else if (ibblig
== 2 && i%ntwin ==0) then // identify the bound
ligand according to MSD
For ilig
= 1, …, nlig
dlig = msd(atom_l,
ntmsd, nftau)
If (dmin <= dlig) blig_min=ilig; dmin=dlig
End
If (dmin
<= dblig) blig=blig_min
End if
If (blig != lig0) Swap atomic coordinates, forces and
velocities of ligand blig with lig0 for
selective higher boost
deltaV =
0.5*k0*(E-V)**2/(Vmax-Vmin)
V = V + deltaV
End
}
Pep-GaMD Algorithm
Pep-GaMD {
If (irest_gamd == 0) then
For i
= 1, …, ntcmd // run initial conventional molecular
dynamics
If (i >= ntcmdprep) Update Vmax, Vmin
If (i >= ntcmdprep && i%ntave ==0) Update Vavg, sigmaV
End
Save Vmax,Vmin,Vavg,sigmaV
to “gamd_restart.dat” file
Calc_E_k0(iE,sigma0,Vmax,Vmin,Vavg,sigmaV)
For i = ntcmd+1, …, ntcmd+nteb // Run
biasing molecular dynamics simulation steps
deltaV = 0.5*k0*(E-V)**2/(Vmax-Vmin)
V = V
+ deltaV
If (i >= ntcmd+ntebprep) Update
Vmax, Vmin
If (i >= ntcmd+ntebprep &&
i%ntave ==0) Update Vavg, sigmaV
Calc_E_k0(iE,sigma0,Vmax,Vmin,Vavg,sigmaV)
End
Save Vmax,Vmin,Vavg,sigmaV
to “gamd_restart.dat” file
else if (irest_gamd == 1)
then
Read Vmax,Vmin,Vavg,
sigmaV from “gamd_restart.dat” file
End if
For i = ntcmd+nteb+1, …, nstlim // run production simulation
deltaV =
0.5*k0*(E-V)**2/(Vmax-Vmin)
V = V + deltaV
End
}
PPI-GaMD Algorithm
PPI-GaMD {
If (irest_gamd == 0) then
For i = 1, …, ntcmd // run initial conventional molecular dynamics
If (i >= ntcmdprep) Update Vmax and Vmin of interaction potential energy
If (i >= ntcmdprep && i%ntave ==0) Update Vavg and sigmaV of interaction potential energy
End
Save Vmax,Vmin,Vavg,sigmaV of interaction potential energy to “gamd_restart.dat” file
Calc_E_k0(iE,sigma0,Vmax,Vmin,Vavg,sigmaV)
For i = ntcmd+1, …, ntcmd+nteb // Run biasing molecular dynamics simulation steps
deltaV = 0.5*k0*(E-V)**2/(Vmax-Vmin)
V = V + deltaV
If (i >= ntcmd+ntebprep) Update Vmax and Vmin of interaction potential energy
If (i >= ntcmd+ntebprep && i%ntave ==0) Update Vavg and sigmaV of interaction potential energy
Calc_E_k0(iE,sigma0,Vmax,Vmin,Vavg,sigmaV)
End
Save Vmax,Vmin,Vavg and sigmaV of of interaction potential energy to “gamd_restart.dat” file
else if (irest_gamd == 1) then
Read Vmax,Vmin,Vavg and sigmaV of interaction potential energy from “gamd_restart.dat” file
End if
For i = ntcmd+nteb+1, …, nstlim // run production simulation
deltaV = 0.5*k0*(E-V)**2/(Vmax-Vmin)
V = V + deltaV
End
}
Simulation Parameter Settings
igamd Flag to apply boost potential
= 0 (default) no boost is applied
= 1 boost on the total potential energy only
= 2 boost on the dihedral energy only
= 3 dual boost on both dihedral and total potential energy
= 4 boost on the non-bonded potential energy only
= 5 dual boost on both dihedral and non-bonded potential energy
= 10 boost on non-bonded potential energy of selected region (defined by timask1 and scmask1) as for a ligand (LiGaMD)
= 11 dual boost on both non-bonded potential energy of the bound ligand and remaining potential energy of the rest of the system (LiGaMD_Dual)
= 14 boost on the total potential energy of selected region (defined by timask1 and scmask1) as for a peptide (Pep-GaMD)
= 15 dual boost on both the peptide potential energy and the total system potential energy other than the peptide potential energy (Pep-GaMD_Dual)
iE Flag to set the threshold energy E for applying all boost potentials
= 1 (default) set the threshold energy to the lower bound E = Vmax
= 2 set the threshold energy to the upper bound E = Vmin + (Vmax – Vmin)/k0
iEP Flag to overwrite iE and set the threshold energy E for applying the first boost potential in dual-boost schemes
= 1 (default) set the threshold energy to the lower bound E = Vmax
= 2 set the threshold energy to the upper bound E = Vmin + (Vmax – Vmin)/k0
iED Flag to overwrite iE and set the threshold energy E for applying the second boost potential in dual-boost schemes
= 1 (default) set the threshold energy to the lower bound E = Vmax
= 2 set the threshold energy to the upper bound E = Vmin + (Vmax – Vmin)/k0
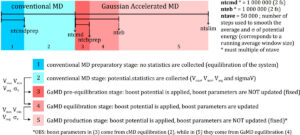
ntcmdprep The number of preparation conventional molecular dynamics steps. This is used for system equilibration and the potential energies are not collected for calculating their statistics. The default is 200,000 for a simulation with 2 fs timestep.
ntcmd The number of initial conventional molecular dynamics simulation steps. Potential energies are collected between ntcmdprep and ntcmd to calculate their maximum, minimum, average and standard deviation (Vmax, Vmin, Vavg, sigmaV). The default is 1,000,000 for a simulation with 2 fs timestep.
ntebprep The number of preparation biasing molecular dynamics simulation steps. This is used for system equilibration after adding the boost potential and the potential statistics (Vmax, Vmin, Vavg, sigmaV) are not updated during these steps. The default is 200,000 for a simulation with 2 fs timestep.
nteb The number of biasing molecular dynamics simulation steps. Potential statistics (Vmax, Vmin, Vavg, sigmaV) are updated between the ntebprep and nteb steps and used to calculate the GaMD acceleration parameters, particularly E and k0. The default is 1,000,000 for a simulation with 2 fs timestep. A greater value may be needed to ensure that the potential statistics and GaMD acceleration parameters level off before running production simulation between the nteb and nstlim (total simulation length) steps. Moreover, nteb can be set to nstlim, by which the potential statistics and GaMD acceleration parameters are updated adaptively throughout the simulation. This in some cases provides more appropriate acceleration.
ntave The number of simulation steps used to calculate the average and standard deviation of potential energies. This variable has already been used in Amber. The default is set to 50,000 for GaMD simulations. It is recommended to be updated as about 4 times of the total number of atoms in the system. Note that ntcmd and nteb need to be multiples of ntave.
irest_gamd Flag to restart GaMD simulation
= 0 (default) new simulation. A file “gamd-restart.dat” that stores the maximum, minimum, average and standard deviation of the potential energies needed to calculate the boost potentials (depending on the igamd flag) will be saved automatically after GaMD equilibration stage.
= 1 restart simulation (ntcmd and nteb are set to 0 in this case). The “gamd-restart.dat” file will be read for restart.
sigma0P The upper limit of the standard deviation of the first potential boost that allows for accurate reweighting. The default is 6.0 (unit: kcal/mol).
sigma0D The upper limit of the standard deviation of the second potential boost that allows for accurate reweighting in dual-boost simulations (e.g., igamd = 2, 3, 5, 11 and 15). The default is 6.0 (unit: kcal/mol).
timask1 Specifies atoms of the first (bound) ligand or peptide in ambmask format when igamd = 10, 11, 14 or 15. The default is an empty string.
scmask1 Specifies atoms of the first (bound) ligand that will be described using soft core in ambmask format in LiGaMD when igamd = 10 or 11. In Pep-GaMD with igamd = 14 or 15, this flag was only used to specify atoms of peptide in ambmask format, but the peptide atoms will not be described using soft core. The default is an empty string.
nlig The total number of ligand molecules in the system. The default is 0. For LiGaMD simulations with nlig>1, it is important to note that: (1) To prepare the simulation starting structure, put the bound ligand first in the PDB, followed by the unbound ligand molecules. (2) Use residue ID of the first (bound) ligand in timask1 and scmask1, for which higher boost potential will be selectively applied. More flexible settings will be provided in future developments of the code.
ibblig The flag to boost the bound ligand selectively with nlig > 1
= 0 (default) no selective boost
= 1 boost the bound ligand selectively out of nlig ligand molecules in the system
= 2 boost the bound ligand selectively out of nlig ligand molecules in the system based on mean-square displacements (MSD)
dblig (Optional) The cutoff distance between atoms atom_p and atom_l used to identify the ligand that is bound in the protein when ibblig = 1 or the cutoff MSD of atom atom_l used to identify the ligand that is bound in the protein when ibblig = 2. If dblig (default 1.0d99 angstroms) is not set in the input file, the first boost potential will be selectively applied to the ligand with the smallest distance to the protein (ibblig = 1) or the smallest MSD (ibblig = 2) out of nlig ligand molecules in the system.
atom_p Serial number of a protein atom (starting from 1 for the first protein atom) used to calculate the ligand distance. It is used only when ibblig = 1. The default is 0.
atom_l Serial number of a ligand atom (starting from 1 for the first ligand atom) used to calculate the ligand distance to the protein. It is used only when ibblig = 1 or 2. The default is 0.
ntmsd Number of timesteps corresponding to the lagging time used to calculate the ligand MSD. It is used only when ibblig = 2. The default is 50000.
nftau Number of saved simulation frames used to calculate the ligand MSD. MSD is calculated for every time window of ntwin = ntmsd + nftau*ntwx steps, for which simulation frames are saved every ntwx steps. It is used only when ibblig = 2. The default is 10.
bgpro2atm Start atomic number of the second protein.
edpro2atm The final atomic number of the second protein.
Known Problems & Solutions
* For new GaMD simulation, there appears abrupt change in the dihedral and total potential energies when “irest=1” (reading both coordinates and velocities) is used. This leads to unexpected large boost potential (last two columns in gamd.log file), for which the normal values should be below 50 kcal/mol.
Solution: This bug has been fixed in AMBER 20.