QAQC Platforms
 The HTSF uses several different platforms to complete quality control testing for samples submitted to our facilities. All material arriving at the HTSF will have QAQC assessments run. Any material that does not meet the requirements for procedures request, will have QAQC sent to the submitter with an explanation as to the issues HTSF see and a discussion possible resolutions.
The HTSF uses several different platforms to complete quality control testing for samples submitted to our facilities. All material arriving at the HTSF will have QAQC assessments run. Any material that does not meet the requirements for procedures request, will have QAQC sent to the submitter with an explanation as to the issues HTSF see and a discussion possible resolutions.
The HTSF does NOT use a Nanodrop for QAQC. Nanodrop is a spectrophotometer, also known as UV-Vis spectrophotometry. While it is useful to determine a sample’s purity (A230:A260 and A260:A280), it is designed to measure a sample’s relative abundance of nucleotides. The Nanodrop does not measure the amount of sample that is intact double stranded, nicked or single stranded DNA/RNA, which is what a fluorometer, such as the Qubit, measures. The Nanodrop is known for incorrect values due to user error and can have measurements 10x different from more stringent methods.
Qubit

Our Qubit Fluorometers are designed to precisely measure the concentration and presence of DNA in an entire sample. It is the most accurate way to measure the concentration of DNA; however, it does not designate DNA fragments. For example, it is unable to determine that there is an overabundance of excessively large or small fragments.
qPCR
The HTSF is able to run qPCR on samples submitted to our facility. Quantitative PCR works to calculate a concentration by amplifying and fluorescently tagging each DNA strand and measuring the change in tagged molecules after each cycle. When coupled with fragment size via Tapestation or Labchip, the HTSF is able to calculate a pool or library’s molarity.Tapestation
 Our Agilent Tapestation machine measures what is actually present in the sample. It shows both the fragment size and the concentration in the given range measured. A graph computed by the Tapestation shows concentration of certain fragments by calculating the area under the curve. Examples of Tapestation results are shown below.
Our Agilent Tapestation machine measures what is actually present in the sample. It shows both the fragment size and the concentration in the given range measured. A graph computed by the Tapestation shows concentration of certain fragments by calculating the area under the curve. Examples of Tapestation results are shown below.
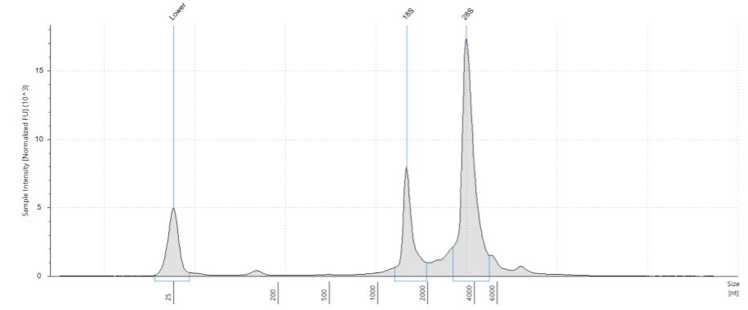
Figure 1: Optimal RNA QAQC Results from Tapestration
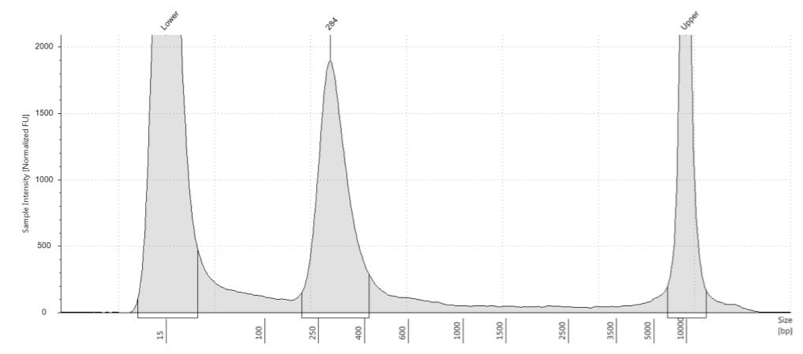
Figure 2: Optimal Library QAQC Results from Tapestration
LabChip

Our LabChip machine is similar to the Tapestation, also determining fragment sizes and concentration. It is used when we are QAQCing many samples at once. Examples of LabChip results are shown below.
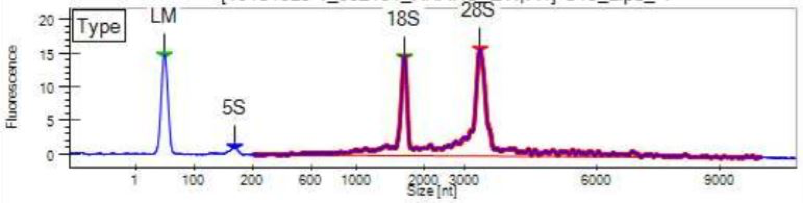
Figure 3: Optimal RNA QAQC Results from LabChip
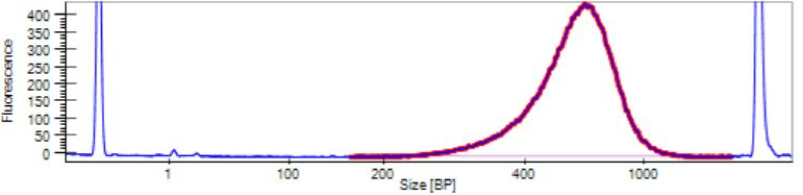
Figure 4: Optimal Library QAQC Results from LabChip