Congratulations to first author Venkat R Chirasani for his research “Whole proteome mapping of compound-protein interactions” published in Current Research in Chemical Biology.
Abstract
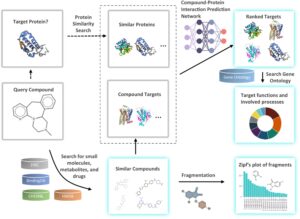 Off-target binding is one of the primary causes of toxic side effects of drugs in clinical development, resulting in failures of clinical trials. While off-target drug binding is a known phenomenon, experimental identification of the undesired protein binders can be prohibitively expensive due to the large pool of possible biological targets. Here, we propose a new strategy combining chemical similarity principle and deep learning to enable proteome-wide mapping of compound-protein interactions. We have developed a pipeline to identify the targets of bioactive molecules by matching them with chemically similar annotated “bait” compounds and ranking them with deep learning. We have constructed a user-friendly web server for drug-target identification based on chemical similarity (DRIFT) to perform searches across annotated bioactive compound datasets, thus enabling high-throughput, multi-ligand target identification, as well as chemical fragmentation of target-binding moieties.
Off-target binding is one of the primary causes of toxic side effects of drugs in clinical development, resulting in failures of clinical trials. While off-target drug binding is a known phenomenon, experimental identification of the undesired protein binders can be prohibitively expensive due to the large pool of possible biological targets. Here, we propose a new strategy combining chemical similarity principle and deep learning to enable proteome-wide mapping of compound-protein interactions. We have developed a pipeline to identify the targets of bioactive molecules by matching them with chemically similar annotated “bait” compounds and ranking them with deep learning. We have constructed a user-friendly web server for drug-target identification based on chemical similarity (DRIFT) to perform searches across annotated bioactive compound datasets, thus enabling high-throughput, multi-ligand target identification, as well as chemical fragmentation of target-binding moieties.
Title: Whole proteome mapping of compound-protein interactions
First author: Venkat R Chirasani, Research Associate
This article was accepted on September 9, 2022.
Link to publication https://doi.org/10.1016/j.crchbi.2022.100035
